%%html
<script src="https://bits.csb.pitt.edu/preamble.js"></script>
import re
m = re.search(r'([^:]):(\d+)([^:]?):([^:]+)(:([^:]+))?',"A:44B:CYS:SG")
%%html
<div id="regexqq" style="width: 500px"></div>
<script>
var divid = '#regexqq';
jQuery(divid).asker({
id: divid,
question: "What is m.group(3)?",
answers: ['Error','SG','CYS','44B','44','44B',':SG'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
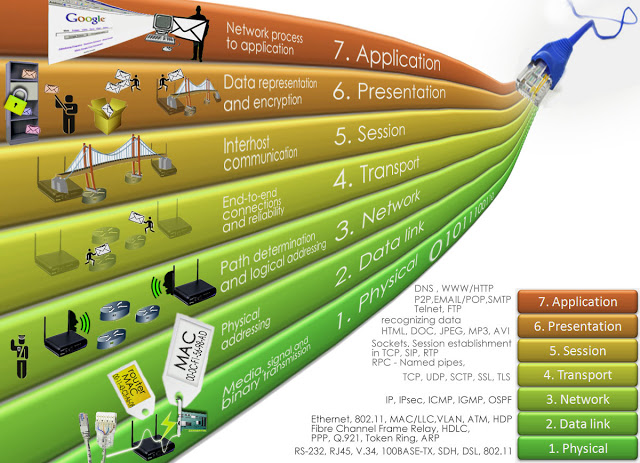
HTTP¶
Hypertext Transfer Protocol
A request-response protocol in a client-server framework.
HTTP Requests¶
The request consists of the following:
- A request line with desired method (action)
- Request Headers
- An empty line.
- An optional message body.
Example
GET / HTTP/1.1 Host: cnn.com Connection: keep-alive Accept: text/html,application/xhtml+xml,application/xml;q=0.9,image/webp,*/*;q=0.8 User-Agent: Mozilla/5.0 (Macintosh; Intel Mac OS X 10_8_5) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/30.0.1599.69 Safari/537.36 Accept-Encoding: gzip,deflate,sdch Accept-Language: en-US,en;q=0.8 Cookie: SelectedEdition=www; optimizelyEndUserId=oeu1364949768474r0.014468349516391754; optimizelySegments=%7B%22170962340%22%3A%22false%22%2C%22171657961%22%3A%22gc%22%2C%22172148679%22%3A%22none%22%2C%22172265329%22%3A%22search%22%7D; optimizelyBuckets=%7B%7D; s_vi=[CS]v1|25F54343051D3284-6000013900246AA6[CE]
HTTP Requests¶
GET
Requests a representation of the specified resource. Requests using GET should only retrieve data and should have no other effect.
POST
Submits data to the server in the request body. Can have side-effects.
HEAD
Asks for the response identical to the one that would correspond to a GET request, but without the response body (just the headers).
OPTIONS, PUT, DELETE, TRACE and CONNECT
HTTP 1.1 methods that are less commonly used.
Sending Data¶
GET
Data must be in the query string of the URL. This is the part of the URL after a question mark:
http://server/program/path/?query_stringThe query_string is made up of name=value pairs separated by &:
http://server/program/path/?field1=value1&field2=value2&field3=value3URLs have length limits (differs by browser, but generally needs to be <2000 characters)
POST
The data is sent in the request body. There is no length limit (just patience limit).
%%html
<div id="httpgetdata" style="width: 500px"></div>
<script>
var divid = '#httpgetdata';
jQuery(divid).asker({
id: divid,
question: "How many data items are being sent in this URL?",
answers: ['0','1','2','3','4'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
HTTP Response¶
A response consists of the following:
- A Status-Line (includes response code)
- Response Headers
- An empty line
- An optional message body
Example:
HTTP/1.1 200 OK
Server: nginx
Date: Thu, 10 Oct 2013 14:23:59 GMT
Content-Type: text/html
Transfer-Encoding: chunked
Connection: keep-alive
Set-Cookie: CG=US:PA:Pittsburgh; path=/
Last-Modified: Thu, 10 Oct 2013 14:23:14 GMT
Vary: Accept-Encoding
Cache-Control: max-age=60, private
Expires: Thu, 10 Oct 2013 14:24:58 GMT
Content-Encoding: gzip
<!DOCTYPE HTML> <html lang="en-US"> <head> <title>CNN.com - Breaking News, U.S., World, Weather, Entertainment & Video News</title>
requests¶
requests is a simple but high-level interface for requesting http data
Also:
urllib2- another high-level interfaceurllib- similar tourllib2but different, hasurlencodefunctionurllib3- successor tourllib2but differenthttplib- low-level interface to http requestmechanize- much higher level interface for scripting web interactions - use this if you need to submit form data, passwords, etc.
get¶
requests.get takes a URL and returns a response object that contains the message body.
Note: urllib2 and mechanize have a urlopen method that returns a file-like object.
import requests
response = requests.get('http://mscbio2025.csb.pitt.edu')
print(response.text)
<!DOCTYPE html>
<html>
<head>
<meta http-equiv="refresh" content="0; url='https://mscbio2025.github.io/'" />
</head>
<body>
</body>
</html>
Must include protocol in URL
fail = requests.get('www.cnn.com')
--------------------------------------------------------------------------- MissingSchema Traceback (most recent call last) Cell In[4], line 1 ----> 1 fail = requests.get('www.cnn.com') File ~/.local/lib/python3.10/site-packages/requests/api.py:73, in get(url, params, **kwargs) 62 def get(url, params=None, **kwargs): 63 r"""Sends a GET request. 64 65 :param url: URL for the new :class:`Request` object. (...) 70 :rtype: requests.Response 71 """ ---> 73 return request("get", url, params=params, **kwargs) File ~/.local/lib/python3.10/site-packages/requests/api.py:59, in request(method, url, **kwargs) 55 # By using the 'with' statement we are sure the session is closed, thus we 56 # avoid leaving sockets open which can trigger a ResourceWarning in some 57 # cases, and look like a memory leak in others. 58 with sessions.Session() as session: ---> 59 return session.request(method=method, url=url, **kwargs) File ~/.local/lib/python3.10/site-packages/requests/sessions.py:575, in Session.request(self, method, url, params, data, headers, cookies, files, auth, timeout, allow_redirects, proxies, hooks, stream, verify, cert, json) 562 # Create the Request. 563 req = Request( 564 method=method.upper(), 565 url=url, (...) 573 hooks=hooks, 574 ) --> 575 prep = self.prepare_request(req) 577 proxies = proxies or {} 579 settings = self.merge_environment_settings( 580 prep.url, proxies, stream, verify, cert 581 ) File ~/.local/lib/python3.10/site-packages/requests/sessions.py:486, in Session.prepare_request(self, request) 483 auth = get_netrc_auth(request.url) 485 p = PreparedRequest() --> 486 p.prepare( 487 method=request.method.upper(), 488 url=request.url, 489 files=request.files, 490 data=request.data, 491 json=request.json, 492 headers=merge_setting( 493 request.headers, self.headers, dict_class=CaseInsensitiveDict 494 ), 495 params=merge_setting(request.params, self.params), 496 auth=merge_setting(auth, self.auth), 497 cookies=merged_cookies, 498 hooks=merge_hooks(request.hooks, self.hooks), 499 ) 500 return p File ~/.local/lib/python3.10/site-packages/requests/models.py:368, in PreparedRequest.prepare(self, method, url, headers, files, data, params, auth, cookies, hooks, json) 365 """Prepares the entire request with the given parameters.""" 367 self.prepare_method(method) --> 368 self.prepare_url(url, params) 369 self.prepare_headers(headers) 370 self.prepare_cookies(cookies) File ~/.local/lib/python3.10/site-packages/requests/models.py:439, in PreparedRequest.prepare_url(self, url, params) 436 raise InvalidURL(*e.args) 438 if not scheme: --> 439 raise MissingSchema( 440 f"Invalid URL {url!r}: No scheme supplied. " 441 f"Perhaps you meant https://{url}?" 442 ) 444 if not host: 445 raise InvalidURL(f"Invalid URL {url!r}: No host supplied") MissingSchema: Invalid URL 'www.cnn.com': No scheme supplied. Perhaps you meant https://www.cnn.com?
Requesting with Data (POST)¶
url = 'http://pocketquery.csb.pitt.edu/pocket.cgi'
values = {'json' : '{"pdbid_text":["1ycr"],"start":0,"num":24,"sort":"score","dir":"desc","cmd":"cluster"}'}
data = requests.post(url,data=values)
print(data)
<Response [200]>
the_page = data.text
the_page.split('\n')[16:18]
['1YCR,B,178,2,5.856,19;23,PHE;TRP,-6.31,-12.62,-6.61,-6.01,5.20125,10.4025,4.7898,5.6127,133.9,267.8,131.1,136.7,72.3,144.6,64.7,79.9,0,0,0,0,0,0,0,0,0.972816', '1YCR,B,129,1,0,19,PHE,-6.61,-6.61,-6.61,-6.61,5.6127,5.6127,5.6127,5.6127,131.1,131.1,131.1,131.1,79.9,79.9,79.9,79.9,0,0,0,0,0,0,0,0,0.96009']
Requesting with Data (GET)¶
values = {'structureId' : '1ycr'}
data = requests.get('http://www.pdb.org/pdb/explore/explore.do',values)
print(data.text)
<!DOCTYPE html><html lang="en"><head><script src="https://www.googletagmanager.com/gtag/js?id=G-5JMGYPWJRR" async></script><script>//- global rcsb-config object
var RC = {
googleAnalyticsTrackingId: 'UA-3923365-3'
, instance: 'production'
, isProductionServer: true
, dataUrl: 'https://data.rcsb.org/'
, searchUrl: 'https://search.rcsb.org/rcsbsearch/v2/'
, alignmentUrl: 'https://alignment.rcsb.org/api/v1-beta/'
, internalAnalyticsOriginHeaderKey: 'Rcsb-Analytics-Traffic-Origin'
, internalAnalyticsOriginHeaderValue: 'internal'
, internalAnalyticsStageHeaderKey: 'Rcsb-Analytics-Traffic-Stage'
, internalAnalyticsStageHeaderValue: 'production'
, MOLSTAR_IMG_URL: 'https://cdn.rcsb.org/images/structures/'
};
</script><script src="/search/search-data?ts=5660297"></script><script src="/js/search/react-search.js?ts=5660297"></script><script>!function(){if("performance"in window==0&&(window.performance={}),Date.now=Date.now||function(){return(new Date).getTime()},"now"in window.performance==0){var n=Date.now();performance.timing&&performance.timing.navigationStart&&(n=performance.timing.navigationStart),window.performance.now=function(){return Date.now()-n}}}();(function(){var h="undefined"!=typeof window&&window===this?this:"undefined"!=typeof global&&null!=global?global:this,k="function"==typeof Object.defineProperties?Object.defineProperty:function(a,b,c){a!=Array.prototype&&a!=Object.prototype&&(a[b]=c.value)};function l(){l=function(){};h.Symbol||(h.Symbol=m)}var n=0;function m(a){return"jscomp_symbol_"+(a||"")+n++}
function p(){l();var a=h.Symbol.iterator;a||(a=h.Symbol.iterator=h.Symbol("iterator"));"function"!=typeof Array.prototype[a]&&k(Array.prototype,a,{configurable:!0,writable:!0,value:function(){return q(this)}});p=function(){}}function q(a){var b=0;return r(function(){return b<a.length?{done:!1,value:a[b++]}:{done:!0}})}function r(a){p();a={next:a};a[h.Symbol.iterator]=function(){return this};return a}function t(a){p();var b=a[Symbol.iterator];return b?b.call(a):q(a)}
function u(a){if(!(a instanceof Array)){a=t(a);for(var b,c=[];!(b=a.next()).done;)c.push(b.value);a=c}return a}var v=0;function w(a,b){var c=XMLHttpRequest.prototype.send,d=v++;XMLHttpRequest.prototype.send=function(f){for(var e=[],g=0;g<arguments.length;++g)e[g-0]=arguments[g];var E=this;a(d);this.addEventListener("readystatechange",function(){4===E.readyState&&b(d)});return c.apply(this,e)}}
function x(a,b){var c=fetch;fetch=function(d){for(var f=[],e=0;e<arguments.length;++e)f[e-0]=arguments[e];return new Promise(function(d,e){var g=v++;a(g);c.apply(null,[].concat(u(f))).then(function(a){b(g);d(a)},function(a){b(a);e(a)})})}}var y="img script iframe link audio video source".split(" ");function z(a,b){a=t(a);for(var c=a.next();!c.done;c=a.next())if(c=c.value,b.includes(c.nodeName.toLowerCase())||z(c.children,b))return!0;return!1}
function A(a){var b=new MutationObserver(function(c){c=t(c);for(var b=c.next();!b.done;b=c.next())b=b.value,"childList"==b.type&&z(b.addedNodes,y)?a(b):"attributes"==b.type&&y.includes(b.target.tagName.toLowerCase())&&a(b)});b.observe(document,{attributes:!0,childList:!0,subtree:!0,attributeFilter:["href","src"]});return b}
function B(a,b){if(2<a.length)return performance.now();var c=[];b=t(b);for(var d=b.next();!d.done;d=b.next())d=d.value,c.push({timestamp:d.start,type:"requestStart"}),c.push({timestamp:d.end,type:"requestEnd"});b=t(a);for(d=b.next();!d.done;d=b.next())c.push({timestamp:d.value,type:"requestStart"});c.sort(function(a,b){return a.timestamp-b.timestamp});a=a.length;for(b=c.length-1;0<=b;b--)switch(d=c[b],d.type){case "requestStart":a--;break;case "requestEnd":a++;if(2<a)return d.timestamp;break;default:throw Error("Internal Error: This should never happen");
}return 0}function C(a){a=a?a:{};this.w=!!a.useMutationObserver;this.u=a.minValue||null;a=window.__tti&&window.__tti.e;var b=window.__tti&&window.__tti.o;this.a=a?a.map(function(a){return{start:a.startTime,end:a.startTime+a.duration}}):[];b&&b.disconnect();this.b=[];this.f=new Map;this.j=null;this.v=-Infinity;this.i=!1;this.h=this.c=this.s=null;w(this.m.bind(this),this.l.bind(this));x(this.m.bind(this),this.l.bind(this));D(this);this.w&&(this.h=A(this.B.bind(this)))}
C.prototype.getFirstConsistentlyInteractive=function(){var a=this;return new Promise(function(b){a.s=b;"complete"==document.readyState?F(a):window.addEventListener("load",function(){F(a)})})};function F(a){a.i=!0;var b=0<a.a.length?a.a[a.a.length-1].end:0,c=B(a.g,a.b);G(a,Math.max(c+5E3,b))}
function G(a,b){!a.i||a.v>b||(clearTimeout(a.j),a.j=setTimeout(function(){var b=performance.timing.navigationStart,d=B(a.g,a.b),b=(window.a&&window.a.A?1E3*window.a.A().C-b:0)||performance.timing.domContentLoadedEventEnd-b;if(a.u)var f=a.u;else performance.timing.domContentLoadedEventEnd?(f=performance.timing,f=f.domContentLoadedEventEnd-f.navigationStart):f=null;var e=performance.now();null===f&&G(a,Math.max(d+5E3,e+1E3));var g=a.a;5E3>e-d?d=null:(d=g.length?g[g.length-1].end:b,d=5E3>e-d?null:Math.max(d,
f));d&&(a.s(d),clearTimeout(a.j),a.i=!1,a.c&&a.c.disconnect(),a.h&&a.h.disconnect());G(a,performance.now()+1E3)},b-performance.now()),a.v=b)}
function D(a){a.c=new PerformanceObserver(function(b){b=t(b.getEntries());for(var c=b.next();!c.done;c=b.next())if(c=c.value,"resource"===c.entryType&&(a.b.push({start:c.fetchStart,end:c.responseEnd}),G(a,B(a.g,a.b)+5E3)),"longtask"===c.entryType){var d=c.startTime+c.duration;a.a.push({start:c.startTime,end:d});G(a,d+5E3)}});a.c.observe({entryTypes:["longtask","resource"]})}C.prototype.m=function(a){this.f.set(a,performance.now())};C.prototype.l=function(a){this.f.delete(a)};
C.prototype.B=function(){G(this,performance.now()+5E3)};h.Object.defineProperties(C.prototype,{g:{configurable:!0,enumerable:!0,get:function(){return[].concat(u(this.f.values()))}}});var H={getFirstConsistentlyInteractive:function(a){a=a?a:{};return"PerformanceLongTaskTiming"in window?(new C(a)).getFirstConsistentlyInteractive():Promise.resolve(null)}};
"undefined"!=typeof module&&module.exports?module.exports=H:"function"===typeof define&&define.amd?define("ttiPolyfill",[],function(){return H}):window.ttiPolyfill=H;})();function gtag(){dataLayer.push(arguments)}var logger=function(){"use strict";function n(n){if(n=JSON.stringify(n),"sendBeacon"in navigator)navigator.sendBeacon(t,n);else{var e=new XMLHttpRequest;e.open("POST",t,!1),e.setRequestHeader("Content-Type","text/plain;charset=UTF-8"),e.send(n)}}function e(e,t,i){if(!RC.isProductionServer)switch(e){case"error":case"warn":case"info":i?console[e](t,i):console[e](t);break;default:console.log(t,i)}"error"!==e&&"warn"!==e||(r.push({level:e,msg:t||"",info:Object.assign({},i,{path:window.location.pathname,userAgent:navigator.userAgent})}),r.length>=o&&n(r.splice(0,o)))}var t="/analytics",o=10,r=[];return window.addEventListener("unload",function(){n(r)},!1),{stack:Function.prototype.bind.call(function(n){n&&e("error",n.name+': "'+n.message+'"',{type:n.name,description:n.message,trace:n.stack})}),error:Function.prototype.bind.call(function(n,t){e("error",n,t)}),warn:Function.prototype.bind.call(function(n,t){e("warn",n,t)}),info:Function.prototype.bind.call(function(n,t){e("info",n,t)}),verbose:Function.prototype.bind.call(function(n,t){e("verbose",n,t)}),debug:Function.prototype.bind.call(function(n,t){e("debug",n,t)}),silly:Function.prototype.bind.call(function(n,t){e("silly",n,t)})}}();window.dataLayer=window.dataLayer||[],gtag("js",new Date),gtag("config","G-5JMGYPWJRR"),window.addEventListener("error",function(n){logger.stack(n.error)}),window.addEventListener("unhandledrejection",function(n){n&&n.reason&&logger.error('Unhandled promise rejection: "'+n.reason.message+'"',{type:"PromiseRejection",description:n.reason.message,trace:n.reason.stack})});var gtagFirstMeaningfulPaint=function(){var n=!1;return function(){n?logger.warn('Timing for "firstMeaningfulPaint" already sent'):(console.log("firstMeaningfulPaint",Math.round(performance.now())),n=!0)}}();!function(){"use strict";var n=0;if("PerformanceObserver"in window&&"PerformancePaintTiming"in window){new PerformanceObserver(function(e){e.getEntries().forEach(function(e){if(0===n){e.startTime,e.duration}})}).observe({entryTypes:["paint"]});var e=window.__tti={e:[]};e.o=new PerformanceObserver(function(n){e.e=e.e.concat(n.getEntries())}),e.o.observe({entryTypes:["longtask"]}),ttiPolyfill.getFirstConsistentlyInteractive({}).then(function(n){});new PerformanceObserver(function(n){n.getEntries().forEach(function(n){var e=n.attribution[0]||{},t={containerType:e.containerType,containerName:e.containerName,containerId:e.containerId,containerSrc:e.containerSrc,frameOrigin:n.name,durationMs:n.duration,startTimeMs:n.startTime};n.duration>500&&logger.warn("Long running task, duration "+n.duration.toFixed(2)+"ms.",t)})}).observe({entryTypes:["longtask"]})}var t=performance.now();window.addEventListener("visibilitychange",function(){document.hidden?t=performance.now():n+=performance.now()-t}),window.addEventListener("load",function(){performance.now()}),window.addEventListener("DOMContentLoaded",function(){performance.now()})}();</script><title>RCSB PDB - 1YCR: MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53</title><meta http-equiv="Content-Type" content="text/html; charset=UTF-8"><meta property="og:title" content="RCSB PDB - 1YCR: MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53"><meta property="og:description" content="MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53"><meta property="og:image" content="https://cdn.rcsb.org/images/structures/1ycr_assembly-1.jpeg"><meta name="twitter:card" content="summary_large_image"><meta name="twitter:title" content="RCSB PDB - 1YCR: MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53"><meta name="twitter:description" content="MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53"><meta name="twitter:image" content="https://cdn.rcsb.org/images/structures/1ycr_assembly-1.jpeg"><meta name="description" content="MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53"><meta name="author" content="RCSB Protein Data Bank"><meta name="email" content="info@rcsb.org"><meta name="Charset" content="UTF-8"><meta name="Distribution" content="Global"><meta name="Rating" content="General"><meta http-equiv="content-language" content="en-US"><meta name="viewport" content="width=device-width, initial-scale=1"><meta name="theme-color" content="#5e94c3"><meta class="elastic" name="release_date" content="release.date:undef"><link rel="manifest" href="/manifest.json"><link rel="apple-touch-icon" href="/img/rcsb-apple-touch-icon.jpg"><link rel="stylesheet" href="https://cdn.rcsb.org/javascript/bootstrap/latest/css/bootstrap.min.css"><link rel="stylesheet" href="https://cdn.rcsb.org/javascript/fontawesome/latest/css/font-awesome.min.css"><link rel="stylesheet" href="/build/css/main.css"><link rel="stylesheet" href="/css/search.css?ts=5660297"><script>window.addEventListener('load', function () {
// load the JIRA feedback button only after everything else has been loaded
var script = document.createElement("script");
script.src = "/js/jira-fdbck.js";
script.type = "text/javascript";
document.getElementsByTagName("head")[0].appendChild(script);
var link = document.createElement("link");
link.rel = "stylesheet";
link.href = "https://cdn.rcsb.org/jira-feedback/css/jira-fdbck.css";
document.getElementsByTagName("head")[0].appendChild(link);
// load the ekko-lightbox css only after everything else has been loaded
var link = document.createElement("link");
link.rel = "stylesheet";
link.href = "/css/ekko-lightbox.css";
document.getElementsByTagName("head")[0].appendChild(link);
});
</script><script src="https://cdn.rcsb.org/javascript/jquery/jquery-3.3.1.min.js"></script><script src="https://cdn.rcsb.org/javascript/bootstrap/latest/js/bootstrap.min.js"></script><script src="/js/ekko-lightbox.min.js" defer></script><script>$(document).delegate('*[data-toggle="lightbox"]',"click",function(t){t.preventDefault(),$(this).ekkoLightbox()});$(function(){$(".nav>.dropdown").on("mouseenter",function(o){$(this).addClass("open")}),$(".nav>.dropdown").on("mouseleave",function(o){$(".dropdown").removeClass("open")}),$('[data-toggle="tooltip"]').tooltip()});</script><link rel="stylesheet" href="/css/pages/staticpages.css"><link rel="stylesheet" href="/css/structure/structuresummarypage.css"><link rel="stylesheet" href="/css/bootstrap-slider.css"><link rel="stylesheet" href="/css/material-switch.css"><script type="text/javascript" src="/saguaro/app.js"></script><script type="text/javascript" src="/saguaro/plot.js"></script><style>.nav-tabs>li.active>a,.nav-tabs>li.active>a:focus,.nav-tabs>li.active>a:hover{color:#fff;cursor:default;background-color:#5e6973;border:1px solid #ddd;border-bottom-color:transparent}#structuretabs>li.active>a,#structuretabs>li.active>a:focus,#structuretabs>li.active>a:hover{background-color:#325880;border:1px solid transparent;border-bottom:none}#structuretabs{border-bottom:5px solid #325880}#structuretabs>li{margin-bottom:0}#structuretabs>li>a{background-color:#efeded;border-bottom:none}#navbar-collapse-SSP{padding:0;margin:0}.secondaryheader h1{float:left;margin-right:10px}.secondaryheader h4{margin-top:30px}#entitynext{margin-top:10px;background-color:#416d93}.panel-default>.panel-heading{color:#fff;background-color:#5e6973;border-color:#ddd}table .highlight td{background-color:#fff2e0}table td ul{-webkit-padding-start:15px}.noteSearchQuery{margin-bottom:0;padding:5px}.querySearchLink{font-weight:700;text-decoration:underline}table tr td button{margin:4px 1px}table thead .externalannotation>th{background-color:#f0ad4e;color:#fff;border-bottom-width:1px;font-weight:400}#structuretabs .dropdown .dropdown-menu{min-width:100%}.actionButtons{display:flex;justify-content:flex-end;margin-bottom:0}@media screen and (min-width:768px) and (max-width:991px){.actionButtons{margin-top:20px;margin-bottom:20px;justify-content:start}}@media screen and (max-width:767px){#collapsedHeader>div{padding-left:0;padding-right:0}#SSP-collapsed-tabs{background-color:#efeded;margin-top:0;border:1px solid #ccc}#navbar-collapse-SSP{width:100%;margin:0}#structuretabs{border:0}#structuretabs li{width:100%}#collapsedHeader h1{float:left;width:100%}.actionButtons{margin-top:20px;margin-bottom:20px;justify-content:center}}</style><script>var structureId = "1YCR";
console.log("CHECK: Summary Structure ID: " + structureId);
var proteinNumber = 0, nonProteinNumber = 0;
</script><script>proteinNumber = 2 + proteinNumber,
nonProteinNumber = 0 + nonProteinNumber;
</script></head><body><div data-elastic-exclude><div data-elastic-exclude><nav class="navbar navbar-inverse navbar-fixed-top hidden-print" id="nav" role="navigation"><div class="hide hidden-print" id="UnsupportedBrowser" style="width: 100%; border-bottom: 2px solid #FFA500; background-color: #FFFFFF; color: #000000; display: inline-block; font-size: .9em; line-height: 2em; text-align: center; font-weight: bold; font-style: italic;"><span class="label label-warning" style="font-size: 100%;">Warning</span> You are using a web browser that we do not support. Our website will not work properly. Please update to a newer version or download a new web browser, such as Chrome or Firefox.</div><div class="container"><div class="row pull-right"><div class="col-xs-6"><div id="mypdb-menu-container"></div></div><div class="col-xs-6" style="padding-top:10px; margin-left: -15px"> <div class="button btn btn-sm basic jira-fdbck-btn" id="feedbackLink-ContactUs">Contact us</div></div></div><!-- Brand and toggle get grouped for better mobile display--><div class="navbar-header"><button class="navbar-toggle pull-left" type="button" data-toggle="collapse" data-target="#navbar-collapse-RCSB"><span class="sr-only">Toggle navigation</span><span class="icon-bar"></span><span class="icon-bar"></span><span class="icon-bar"></span></button><a class="navbar-brand" href="/">RCSB PDB</a></div><div class="collapse navbar-collapse" id="navbar-collapse-RCSB" style="max-height: 420px;"><ul class="nav navbar-nav pull-left"><li class="dropdown" id="headnav_deposit"><a class="dropdown-toggle" href="#" data-toggle="dropdown" aria-expanded="false">Deposit <b class="caret"></b></a><div class="dropdown-menu multi-column" id="DepositDropdownNav"><div class="row"><div class="col-xs-12 col-sm-2 col-md-2"><ul class="dropdown-menu" id="head-deposit_prepare"><h4>Prepare Data</h4><li> <a href="https://www.wwpdb.org/deposition/preparing-pdbx-mmcif-files" rel="noopener" target="_blank">PDBx/mmCIF file</a></li><li> <a href="https://pdb-extract.wwpdb.org/" rel="noopener" target="_blank">pdb_extract</a></li><li> <a href="http://sf-tool.wwpdb.org/" rel="noopener" target="_blank">SF-Tool</a></li><li> <a href="http://ligand-expo.rcsb.org/" rel="noopener" target="_blank">Ligand Expo</a></li><li> <a href="https://sw-tools.rcsb.org/apps/MAXIT/index.html" rel="noopener" target="_blank">MAXIT</a></li></ul></div><div class="col-xs-12 col-sm-3 col-md-3"><ul class="dropdown-menu" id="head-deposit_validate"><h4>Validate Data</h4><li> <a href="https://validate-rcsb.wwpdb.org" rel="noopener" target="_blank">Validation Server</a></li><li> <a href="http://www.wwpdb.org/validation/onedep-validation-web-service-interface" rel="noopener" target="_blank">Validation API</a></li><li> <a href="http://www.wwpdb.org/documentation/journals" rel="noopener" target="_blank">Information for Journals</a></li><li> <a href="http://www.wwpdb.org/task/validation-task-forces.php" rel="noopener" target="_blank">Validation Task Forces</a></li></ul></div><div class="col-xs-12 col-sm-3 col-md-3"><ul class="dropdown-menu" id="head-deposit_deposit-data"><h4>Deposit Data</h4><li style="word-break: normal"> <a href="https://deposit.wwpdb.org/deposition/" rel="noopener" target="_blank" style="word-break: normal">wwPDB OneDep System</a></li><li style="word-break: normal"> <a href="https://pdb-dev.wwpdb.org/" rel="noopener" target="_blank" style="word-break: normal">PDB-Dev</a></li></ul></div><div class="col-xs-12 col-sm-2 col-md-3"><ul class="dropdown-menu" id="head-deposit_deposit-help"><h4>Help and Resources</h4><li> <a href="http://www.wwpdb.org/deposition/faq" rel="noopener" target="_blank">Deposit FAQ</a></li><li> <a href="http://www.wwpdb.org/validation/2017/FAQs" rel="noopener" target="_blank">Validation FAQ</a></li><li> <a href="http://www.wwpdb.org/deposition/tutorial" rel="noopener" target="_blank">Tutorials</a></li><li> <a href="http://www.wwpdb.org/documentation/policy" rel="noopener" target="_blank">Annotation Policies</a></li><li> <a href="http://www.wwpdb.org/documentation/procedure" rel="noopener" target="_blank">Processing Procedures</a></li><li> <a href="https://mmcif.wwpdb.org/" rel="noopener" target="_blank">PDBx/mmCIF Dictionary</a></li><li> <a href="https://www.wwpdb.org/data/ccd" rel="noopener" target="_blank">Chemical Component Dictionary</a></li><li> <a href="https://www.wwpdb.org/data/bird" rel="noopener" target="_blank">Biologically Interesting Molecule Reference Dictionary (BIRD)</a></li><li> <a href="https://biosync.sbkb.org/" rel="noopener" target="_blank">BioSync/Beamlines/Facilities</a></li><li> <a href="https://sw-tools.rcsb.org/" rel="noopener" target="_blank">Related Tools</a></li></ul></div></div></div></li><li class="dropdown" id="headnav_search"><a class="dropdown-toggle" href="#" data-toggle="dropdown">Search <b class="caret"></b></a><ul class="dropdown-menu"><li><a href="/search/advanced" rel="noopener">Advanced Search</a></li><li><a href="/search/advanced/sequence" rel="noopener">Sequence Similarity Search</a></li><li><a href="/search/advanced/structure" rel="noopener">Structure Similarity Search</a></li><li><a href="/search/advanced/chemical" rel="noopener">Chemical Similarity Search</a></li><li><a href="/chemical-sketch" rel="noopener">Chemical Sketch Tool</a></li><li><a href="/search/browse/atc" rel="noopener">Browse by Annotations</a></li><li><a href="/search?query=%7B%22parameters%22%3A%7B%22attribute%22%3A%22rcsb_accession_info.initial_release_date%22%2C%22operator%22%3A%22range%22%2C%22value%22%3A%7B%22from%22%3A%22now-1w%22%2C%22include_lower%22%3Afalse%2C%22to%22%3A%22now%22%2C%22include_upper%22%3Atrue%7D%7D%7D" rel="noopener">New Entries</a></li><li><a href="/pages/unreleased" rel="noopener">Unreleased Entries</a></li><li><a href="/stats" rel="noopener">PDB Statistics</a></li></ul></li><li class="dropdown" id="headnav_visualize"><a class="dropdown-toggle" href="#" data-toggle="dropdown">Visualize <b class="caret"></b></a><ul class="dropdown-menu"> <li><a href="/3d-view" rel="noopener">Mol* (MolStar)</a></li><li><a href="/docs/sequence-viewers/protein-feature-view" rel="noopener">Protein Feature View</a></li><li><a href="/docs/sequence-viewers/genome-view" rel="noopener">Genome View</a></li></ul></li><li class="dropdown" id="headnav_analyze"><a class="dropdown-toggle" href="#" data-toggle="dropdown">Analyze <b class="caret"></b></a><ul class="dropdown-menu"><li><a href="/alignment" rel="noopener">Pairwise Structure Alignment</a></li><li><a href="/docs/general-help/symmetry-resources-in-the-pdb" rel="noopener">Symmetry Resources in the PDB</a></li><li><a href="https://www.wwpdb.org/validation/validation-reports" rel="noopener" target="_blank">Structure Quality </a></li><li><a href="/docs/grouping-structures/overview-grouping-structures" rel="noopener">Grouping Structures</a></li><li><a href="https://cdn.rcsb.org/rcsb-pdb/pdb50mesh/index.html" rel="noopener" target="_blank">PDB Citation MeSH Network Explorer </a></li><li><a href="/stats" rel="noopener">PDB Statistics</a></li><li><a href="https://www.eppic-web.org/ewui/" rel="noopener" target="_blank">EPPIC Biological Assemblies </a></li><li class="extraList">External Data and Resources </li><li class="extraMargin"><a href="/docs/general-help/data-from-external-resources-integrated-into-rcsb-pdb" rel="noopener">Integrated Resources </a></li><li class="extraMargin"><a href="/docs/additional-resources/databases-and-knowledgebases" rel="noopener">Additional Resources </a></li></ul></li><li class="dropdown hidden-xs hidden-sm" id="headnav_download"><a class="dropdown-toggle" href="#" data-toggle="dropdown">Download <b class="caret"></b></a><ul class="dropdown-menu"> <li><a href="/downloads" rel="noopener">Coordinates and Experimental Data</a></li><li><a href="/downloads/fasta" rel="noopener">Sequences</a></li><li><a href="/downloads/ligands" rel="noopener">Ligands</a></li><li><a href="/docs/programmatic-access/file-download-services" rel="noopener">File Download Services</a></li><li><a href="/docs/programmatic-access/web-services-overview" rel="noopener">Web Services</a></li></ul></li><li class="dropdown" id="headnav_learn"><a class="dropdown-toggle" href="#" data-toggle="dropdown">Learn <b class="caret"></b></a><ul class="dropdown-menu"><h4><a href="http://pdb101.rcsb.org" target="_blank" rel="noopener"><img class="image_submenu" src="https://cdn.rcsb.org/rcsb-pdb/v2/home/images/menuImgs/pdb101Logo.png"></a></h4><li><a href="https://pdb101.rcsb.org/train/training-events" rel="noopener" target="_blank">Training Courses </a></li><li><a href="https://pdb101.rcsb.org/learn/guide-to-understanding-pdb-data/introduction" rel="noopener" target="_blank">Guide to PDB Data </a></li><li><a href="https://pdb101.rcsb.org/motm/motm-by-date" rel="noopener" target="_blank">Molecule of the Month </a></li><li><a href="https://pdb101.rcsb.org/learn/paper-models" rel="noopener" target="_blank">Educational Resources </a></li><li><a href="https://pdb101.rcsb.org/teach/overview" rel="noopener" target="_blank">Curricula </a></li><li><a href="https://pdb101.rcsb.org/browse" rel="noopener" target="_blank">Browse </a></li><li><a href="https://pdb101.rcsb.org/news/2023" rel="noopener" target="_blank">News </a></li><li class="extraList">SciArt Galleries </li><li class="extraMargin"><a href="https://pdb101.rcsb.org/sci-art/geis-archive/gallery-by-name" rel="noopener" target="blank">Irving Geis </a></li><li class="extraMargin"><a href="https://pdb101.rcsb.org/sci-art/goodsell-gallery" rel="noopener" target="blank">David Goodsell </a></li></ul></li><li class="dropdown" id="headnav_more"><a class="dropdown-toggle" href="#" data-toggle="dropdown">About <b class="caret"></b></a><ul class="dropdown-menu"><li><a href="/pages/contactus" rel="noopener">Contact Us</a></li><li><a href="/pages/about-us/index" rel="noopener">About RCSB PDB</a></li><li><a href="/pages/about-us/mission" rel="noopener">Vision and Mission</a></li><li><a href="/pages/policies" rel="noopener">Citation, Usage, Privacy Policies, Logo</a></li><li><a href="/pages/news" rel="noopener">News</a></li><li><a href="/pages/about-us/history" rel="noopener">PDB History</a></li><li><a href="/pages/pdb50" rel="noopener">PDB50</a></li><li><a href="/pages/about-us/pdb-user-community" rel="noopener">User Community</a></li><li><a href="/pages/publications" rel="noopener">Publications</a></li><li><a href="/pages/advisory-committee" rel="noopener">RCSB PDB Advisory Committee</a></li><li><a href="/pages/team" rel="noopener">Team Members</a></li><li><a href="/pages/about-us/deia" rel="noopener">Diversity, Equity, Inclusion, and Access</a></li><li><a href="https://status.rcsb.org/" rel="noopener" target="_blank">Service Status </a></li></ul></li><li class="dropdown" id="headnav_documentation"><a class="dropdown-toggle" href="#" data-toggle="dropdown">Documentation <b class="caret"></b></a><ul class="dropdown-menu"> <li><a class="extraList" href="/docs/general-help/organization-of-3d-structures-in-the-protein-data-bank" rel="noopener" style="padding-left: 0px;">General Help </a></li><li class="extraMargin"><a href="/docs/general-help/organization-of-3d-structures-in-the-protein-data-bank" rel="noopener">3D Structures in the Protein Data Bank </a></li><li class="extraMargin"><a href="/docs/general-help/computed-structure-models-and-rcsborg" rel="noopener">Computed Structure Models (CSM) </a></li><li><a class="extraList" href="/docs/search-and-browse/overview-search-and-browse" rel="noopener" style="padding-left: 0px;">Search and Browse </a></li><li class="extraMargin"><a href="/docs/search-and-browse/basic-search" rel="noopener">Basic Search </a></li><li class="extraMargin"><a href="/docs/search-and-browse/advanced-search/overview-advanced-search" rel="noopener">Advanced Search </a></li><li class="extraMargin"><a href="/docs/search-and-browse/browse-options/overview-browse" rel="noopener">Browse Annotations </a></li><li><a class="extraList" href="/docs/3d-viewers/mol*/getting-started" rel="noopener" style="padding-left: 0px;">Visualize and Analyze </a></li><li class="extraMargin"><a href="/docs/sequence-viewers/genome-view" rel="noopener">Sequence Viewers </a></li><li class="extraMargin"><a href="/docs/tools/pairwise-structure-alignment" rel="noopener">Pairwise Structure Alignment </a></li><li><a href="/docs/exploring-a-3d-structure/overview" rel="noopener">Exploring a 3D Structure</a></li><li><a href="/docs/grouping-structures/overview-grouping-structures" rel="noopener">Grouping Structures</a></li><li><a href="/docs/programmatic-access/file-download-services" rel="noopener">Programmatic Access</a></li><li><a href="/docs/additional-resources/databases-and-knowledgebases" rel="noopener">Additional Resources</a></li><li><a href="/docs/general-help/software-supporters" rel="noopener">Software Supporters</a></li><li><a href="/docs/general-help/deposition-resources" rel="noopener">Deposition Resources</a></li><li><a href="/docs/general-help/website-faq" rel="noopener">FAQs</a></li><li><a href="/docs/general-help/glossary" rel="noopener">Glossary</a></li></ul></li><li><a class="dropdown-toggle" href="/pages/jobs">Careers</a></li><li><a class="dropdown-toggle" href="/news/feature/5e74d55d2d410731e9944f52">COVID-19</a></li><li class="dropdown visible-xs"><div id="socialmedia_section_mobile"><ul class="fa-ul hidden-print"><li><a href="https://www.facebook.com/RCSBPDB" target="_blank" rel="noopener" title="Facebook" data-placement="top" data-toggle="tooltip"><i class="fa fa-facebook-square fa-lg"></i></a></li><li><a href="https://twitter.com/buildmodels" target="_blank" rel="noopener" title="Twitter" data-placement="top" data-toggle="tooltip"><i class="fa fa-twitter fa-lg"></i></a></li><li><a href="https://www.youtube.com/user/RCSBProteinDataBank" target="_blank" rel="noopener" title="YouTube" data-placement="top" data-toggle="tooltip"><i class="fa fa-youtube-play fa-lg"></i></a></li><li><a href="https://github.com/rcsb" target="_blank" rel="noopener" title="Github" data-placement="top" data-toggle="tooltip"><i class="fa fa-github fa-lg"></i></a></li></ul></div></li></ul></div></div></nav></div><style>#PDBmembers li {margin-right: 15px; }
</style><div data-elastic-exclude><div id="header"><div class="container"><div class="row" style="margin-bottom: 0px;"><div class="col-md-6 col-lg-5 hidden-sm hidden-xs" id="logo_container" style="padding-bottom: 15px;"><table class="header-left"><tr><td><a href="/"><img id="rcsblogo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/rcsb_logo.png" alt="RCSB PDB"></a></td><td><table id="header-counts"><tr><td><div class="results-content-type-sm experimental"><span class="fa fa-flask" data-toggle="tooltip" data-placement="right" data-original-title="Experimental PDB Structure"></span></div></td><td><a href="/search?request=%7B%22query%22%3A%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22text%22%2C%22parameters%22%3A%7B%22attribute%22%3A%22rcsb_entry_info.structure_determination_methodology%22%2C%22operator%22%3A%22exact_match%22%2C%22value%22%3A%22experimental%22%7D%7D%5D%2C%22logical_operator%22%3A%22and%22%7D%5D%2C%22logical_operator%22%3A%22and%22%2C%22label%22%3A%22text%22%7D%5D%2C%22logical_operator%22%3A%22and%22%7D%2C%22return_type%22%3A%22entry%22%2C%22request_options%22%3A%7B%22scoring_strategy%22%3A%22combined%22%2C%22results_content_type%22%3A%5B%22experimental%22%5D%7D%7D" style="font-weight: bold; font-size: 110%">210,836</a><span> Structures from the PDB</span></td></tr><tr><td><div class="results-content-type-sm computational"><span class="fa fa-desktop" data-toggle="tooltip" data-placement="right" data-original-title="Computed Structure Model"></span></div></td><td><a href="/search?request=%7B%22query%22%3A%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22group%22%2C%22nodes%22%3A%5B%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22text%22%2C%22parameters%22%3A%7B%22attribute%22%3A%22rcsb_entry_info.structure_determination_methodology%22%2C%22operator%22%3A%22exact_match%22%2C%22value%22%3A%22computational%22%7D%7D%5D%2C%22logical_operator%22%3A%22and%22%7D%5D%2C%22logical_operator%22%3A%22and%22%2C%22label%22%3A%22text%22%7D%5D%2C%22logical_operator%22%3A%22and%22%7D%2C%22return_type%22%3A%22entry%22%2C%22request_options%22%3A%7B%22scoring_strategy%22%3A%22combined%22%2C%22results_content_type%22%3A%5B%22computational%22%5D%7D%7D" style="font-weight: bold; font-size: 110%">1,068,577</a><span> Computed Structure Models (CSM)</span></td></tr></table></td></tr></table></div><div class="col-sm-12 col-md-6 col-lg-7 hidden-print" id="search-bar-container"></div></div></div><div class="logos_bcg"><div class="container"><ul class="hidden-xs hidden-print" id="PDBmembers"><li><span data-toggle="tooltip" data-placement="right" data-original-title="PDB-101: Training and outreach portal of RCSB PDB"><a href="https://pdb101.rcsb.org/"><img id="pdb101logo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/pdb101-truncated.png" alt="PDB-101"></a></span></li><li><span data-toggle="tooltip" data-placement="right" data-original-title="wwPDB: Worldwide Protein Data Bank"><a href="https://www.wwpdb.org/" target="_blank" rel="noopener"><img id="wwpdblogo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/wwpdb-truncated.png" alt="wwPDB"></a></span></li><li><span data-toggle="tooltip" data-placement="right" data-original-title="EMDataResource: 3DEM resources"><a href="https://www.emdataresource.org" target="_blank" rel="noopener"><img id="emdatabanklogo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/emdb-truncated.png" alt="EMDataResource"></a></span></li><li><span data-toggle="tooltip" data-placement="right" data-original-title="NAKB: Nucleic Acid Knowledgebase"><a href="https://nakb.org/" target="_blank" rel="noopener"><img id="nakblogo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/nakb-logo.png" alt="NAKB: Nucleic Acid Knowledgebase"></a></span></li><li><span data-toggle="tooltip" data-placement="right" data-original-title="wwPDB Foundation: Donate to support wwPDB outreach activities"><a href="https://foundation.wwpdb.org/" target="_blank" rel="noopener"><img id="wwpdbfoundationlogo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/foundation-truncated.png" alt="wwPDB Foundation"></a></span></li><li><span data-toggle="tooltip" data-placement="right" data-original-title="PDB-Dev: Prototype Archiving System for Integrative Structures"><a href="https://pdb-dev.wwpdb.org/" target="_blank" rel="noopener"><img id="pdbdevlogo" src="https://cdn.rcsb.org/rcsb-pdb/v2/common/images/pdb-dev-logo.png" alt="PDB-Dev Logo"></a></span></li></ul><div id="socialmedia_section"><ul class="fa-ul hidden-print"><li><a href="https://www.facebook.com/RCSBPDB" target="_blank" rel="noopener" title="Facebook" data-placement="top" data-toggle="tooltip"><i class="fa fa-facebook-square fa-lg"></i></a></li><li><a href="https://twitter.com/buildmodels" target="_blank" rel="noopener" title="Twitter" data-placement="top" data-toggle="tooltip"><i class="fa fa-twitter fa-lg"></i></a></li><li><a href="https://www.youtube.com/user/RCSBProteinDataBank" target="_blank" rel="noopener" title="YouTube" data-placement="top" data-toggle="tooltip"><i class="fa fa-youtube-play fa-lg"></i></a></li><li><a href="https://github.com/rcsb" target="_blank" rel="noopener" title="Github" data-placement="top" data-toggle="tooltip"><i class="fa fa-github fa-lg"></i></a></li></ul></div></div></div></div></div><div class="container" id="maincontentcontainer"><button class="navbar-toggle pull-left" id="SSP-collapsed-tabs" type="button" data-toggle="collapse" data-target="#navbar-collapse-SSP"><span class="glyphicon glyphicon-menu-hamburger" aria-hidden="true"></span> Navigation Tabs</button><div class="collapse navbar-collapse" id="navbar-collapse-SSP"><ul class="nav nav-tabs hidden-print" id="structuretabs" role="tablist"><li id="structuresummarytab" role="presentation"><a href="/structure/1YCR">Structure Summary</a></li><li id="3dviewtab" role="presentation"><a href="/3d-view/1YCR">3D View</a></li><li id="annotationstab" role="presentation"><a href="/annotations/1YCR">Annotations</a></li><li id="MoreOptions-Method" role="presentation"><a href="/experimental/1YCR">Experiment</a></li><li id="sequencetab" role="presentation"><a href="/sequence/1YCR">Sequence</a></li><li id="genometab" role="presentation"><a href="/genome/1YCR">Genome</a></li><li id="versionstab" role="presentation"><a href="/versions/1YCR">Versions</a></li><!--+tab( 'Sequence', '/pdb/explore/remediatedSequence.do?structureId=' + entryId, 'Sequence' )--><!--+tab( 'MoreOptions-Method', '/experimental/' + entryId, 'Experiment' )--></ul></div><br><div class="tab-content"><div class="tab-pane active" id="summary" role="tabpanel"><div class="row"><div class="col-md-8 col-sm-12 col-xs-12 pull-right"><div class="actionButtons btn-toolbar" role="toolbar"><style>#DownloadFilesButton .dropdown-menu li.filedivider:not(:first-child), #DisplayFilesButton .dropdown-menu li.filedivider:not(:first-child) {
height: 1px;
margin: 9px 0;
overflow: hidden;
background-color: #e5e5e5;
}
</style><div class="btn-group" id="ProductPrimaryActions" role="group"><div class="btn-group" id="DisplayFilesButton" role="group"><button class="btn btn-sm btn-primary dropdown-toggle" id="dropdownMenuDisplayFiles" type="button" data-toggle="dropdown" aria-expanded="false"><span class="fa fa-file" aria-hidden="true"></span> Display Files <span class="caret"></span></button><ul class="dropdown-menu pull-right" role="menu" aria-labelledby="dropdownMenuDisplayFiles"><li class="filedivider"></li><li><a target="_blank" rel="noopener" href="/fasta/entry/1YCR/display">FASTA Sequence</a></li><li class="filedivider"></li><li><a target="_blank" rel="noopener" href="//files.rcsb.org/view/1YCR.cif">mmCIF Format</a></li><li><a target="_blank" rel="noopener" href="//files.rcsb.org/header/1YCR.cif">mmCIF Format (Header)</a></li><li class="filedivider"></li><li><a target="_blank" rel="noopener" href="//files.rcsb.org/view/1YCR.pdb">PDB Format</a></li><li><a target="_blank" rel="noopener" href="//files.rcsb.org/header/1YCR.pdb">PDB Format (Header)</a></li></ul></div><div class="btn-group" id="DownloadFilesButton" role="group"><button class="btn btn-sm btn-primary dropdown-toggle" id="dropdownMenuDownloadFiles" type="button" data-toggle="dropdown" aria-expanded="false"><span class="fa fa-arrow-circle-o-down" aria-hidden="true" style="font-size: 14px;"></span> Download Files <span class="caret"></span></button><ul class="dropdown-menu pull-right" role="menu" aria-labelledby="dropdownMenuDownloadFiles"><li class="filedivider"></li><li><a href="/fasta/entry/1YCR">FASTA Sequence</a></li><li class="filedivider"></li><li><a href="//files.rcsb.org/download/1YCR.cif">PDBx/mmCIF Format</a></li><li><a href="//files.rcsb.org/download/1YCR.cif.gz">PDBx/mmCIF Format (gz)</a></li><li class="filedivider"></li><li><a href="//files.rcsb.org/download/1YCR.pdb">PDB Format</a></li><li><a href="//files.rcsb.org/download/1YCR.pdb.gz">PDB Format (gz)</a></li><li class="filedivider"></li><li><a href="//files.rcsb.org/download/1YCR.xml.gz">PDBML/XML Format (gz)</a></li><li class="filedivider"></li><li><a target="_blank" rel="noopener" href="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_full_validation.pdf">Validation Full PDF</a></li><li><a target="_blank" rel="noopener" href="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_validation.xml.gz">Validation (XML - gz)</a></li><li class="filedivider"></li><li><a href="//files.rcsb.org/download/1YCR-assembly1.cif.gz">Biological Assembly 1 (CIF - gz) <span class="fa fa-info-circle" aria-hidden="true" data-toggle="tooltip" data-placement="top" title="Biological assembly files in .cif format."></span></a></li><li><a href="//files.rcsb.org/download/1YCR.pdb1.gz">Biological Assembly 1 (PDB - gz)</a></li></ul></div></div><div class="btn-group" role="group" style="margin-left: 2px;" target="_blank" data-toggle="tooltip" data-placement="top" data-original-title="Get the Data API query used to generate this page"> <a href="https://data.rcsb.org/graphql/index.html?query=query%20structure%20(%24id%3A%20String!)%20%7B%0A%20%20entry(entry_id%3A%24id)%7B%0A%20%20%20%20%20%20rcsb_id%0A%20%20%20%20%20%20entry%20%7B%0A%20%20%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_entry_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20entity_ids%0A%20%20%20%20%20%20%20%20%20%20pubmed_id%0A%20%20%20%20%20%20%20%20%20%20emdb_ids%0A%20%20%20%20%20%20%20%20%20%20assembly_ids%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_associated_holdings%20%7B%0A%20%20%20%20%20%20%20%20%20%20rcsb_repository_holdings_current_entry_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20assembly_ids%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_repository_holdings_current%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20repository_content_types%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_comp_model_provenance%20%7B%0A%20%20%20%20%20%20%20%20source_url%0A%20%20%20%20%20%20%20%20source_pae_url%0A%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20source_db%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_database_status%20%7B%0A%20%20%20%20%20%20%20%20%20%20pdb_format_compatible%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20struct%20%7B%0A%20%20%20%20%20%20%20%20%20%20title%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_ma_qa_metric_global%20%7B%0A%20%20%20%20%20%20%20%20model_id%0A%20%20%20%20%20%20%20%20ma_qa_metric_global%20%7B%0A%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20value%0A%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20description%0A%20%20%20%20%20%20%20%20%20%20type_other_details%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_primary_citation%20%7B%0A%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20pdbx_database_id_DOI%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pubmed%20%7B%0A%20%20%20%20%20%20%20%20rcsb_pubmed_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20pubmed_id%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20rcsb_pubmed_central_id%0A%20%20%20%20%20%20%20%20rcsb_pubmed_doi%0A%20%20%20%20%20%20%20%20rcsb_pubmed_abstract_text%0A%20%20%20%20%20%20%20%20rcsb_pubmed_affiliation_info%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_deposit_group%20%7B%0A%20%20%20%20%20%20%20%20group_id%0A%20%20%20%20%20%20%20%20group_type%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_external_references%20%7B%0A%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20link%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_database_PDB_obs_spr%20%7B%0A%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20replace_pdb_id%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20struct_keywords%20%7B%0A%20%20%20%20%20%20%20%20%20%20pdbx_keywords%0A%20%20%20%20%20%20%20%20%20%20text%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20exptl%20%7B%0A%20%20%20%20%20%20%20%20%20%20method%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20cell%20%7B%0A%20%20%20%20%20%20%20%20%20%20length_a%0A%20%20%20%20%20%20%20%20%20%20length_b%0A%20%20%20%20%20%20%20%20%20%20length_c%0A%20%20%20%20%20%20%20%20%20%20angle_alpha%0A%20%20%20%20%20%20%20%20%20%20angle_beta%0A%20%20%20%20%20%20%20%20%20%20angle_gamma%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20symmetry%20%7B%0A%20%20%20%20%20%20%20%20%20%20space_group_name_H_M%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20software%20%7B%0A%20%20%20%20%20%20%20%20%20%20classification%0A%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_accession_info%20%7B%0A%20%20%20%20%20%20%20%20%20%20deposit_date%0A%20%20%20%20%20%20%20%20%20%20initial_release_date%0A%20%20%20%20%20%20%20%20%20%20major_revision%0A%20%20%20%20%20%20%20%20%20%20minor_revision%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_audit_revision_history%20%7B%0A%20%20%20%20%20%20%20%20%20%20ordinal%0A%20%20%20%20%20%20%20%20%20%20data_content_type%0A%20%20%20%20%20%20%20%20%20%20major_revision%0A%20%20%20%20%20%20%20%20%20%20minor_revision%0A%20%20%20%20%20%20%20%20%20%20revision_date%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_audit_revision_details%20%7B%0A%20%20%20%20%20%20%20%20ordinal%0A%20%20%20%20%20%20%20%20revision_ordinal%0A%20%20%20%20%20%20%20%20data_content_type%0A%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20description%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_audit_revision_group%20%7B%0A%20%20%20%20%20%20%20%20%20%20ordinal%0A%20%20%20%20%20%20%20%20%20%20revision_ordinal%0A%20%20%20%20%20%20%20%20%20%20data_content_type%0A%20%20%20%20%20%20%20%20%20%20group%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_database_related%20%7B%0A%20%20%20%20%20%20%20%20content_type%0A%20%20%20%20%20%20%20%20db_id%0A%20%20%20%20%20%20%20%20details%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20audit_author%20%7B%0A%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_audit_support%20%7B%0A%20%20%20%20%20%20%20%20funding_organization%0A%20%20%20%20%20%20%20%20country%0A%20%20%20%20%20%20%20%20grant_number%0A%20%20%20%20%20%20%20%20ordinal%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20refine%20%7B%0A%20%20%20%20%20%20%20%20%20%20pdbx_refine_id%0A%20%20%20%20%20%20%20%20%20%20ls_d_res_high%0A%20%20%20%20%20%20%20%20%20%20ls_R_factor_R_work%0A%20%20%20%20%20%20%20%20%20%20ls_R_factor_R_free%0A%20%20%20%20%20%20%20%20%20%20ls_R_factor_obs%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_nmr_ensemble%20%7B%0A%20%20%20%20%20%20%20%20%20%20conformers_calculated_total_number%0A%20%20%20%20%20%20%20%20%20%20conformers_submitted_total_number%0A%20%20%20%20%20%20%20%20%20%20conformer_selection_criteria%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20em_experiment%20%7B%0A%20%20%20%20%20%20%20%20%20%20aggregation_state%0A%20%20%20%20%20%20%20%20%20%20reconstruction_method%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20em_3d_reconstruction%20%7B%0A%20%20%20%20%20%20%20%20%20%20resolution%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20em_software%20%7B%0A%20%20%20%20%20%20%20%20category%20%20%0A%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20version%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20citation%20%7B%0A%20%20%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20%20%20title%0A%20%20%20%20%20%20%20%20%20%20rcsb_journal_abbrev%0A%20%20%20%20%20%20%20%20%20%20rcsb_authors%0A%20%20%20%20%20%20%20%20%20%20year%0A%20%20%20%20%20%20%20%20%20%20journal_volume%0A%20%20%20%20%20%20%20%20%20%20page_first%0A%20%20%20%20%20%20%20%20%20%20page_last%0A%20%20%20%20%20%20%20%20%20%20pdbx_database_id_PubMed%0A%20%20%20%20%20%20%20%20%20%20pdbx_database_id_DOI%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20pdbx_database_related%20%7B%0A%20%20%20%20%20%20%20%20%20%20db_id%0A%20%20%20%20%20%20%20%20%20%20db_name%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_entry_info%20%7B%0A%20%20%20%20%20%20%20%20molecular_weight%0A%20%20%20%20%20%20%20%20deposited_atom_count%0A%20%20%20%20%20%20%20%20deposited_model_count%0A%20%20%20%20%20%20%20%20deposited_polymer_monomer_count%0A%20%20%20%20%20%20%20%20deposited_modeled_polymer_monomer_count%0A%20%20%20%20%20%20%20%20deposited_unmodeled_polymer_monomer_count%0A%20%20%20%20%20%20%20%20polymer_entity_count_protein%0A%20%20%20%20%20%20%20%20polymer_entity_count_nucleic_acid%0A%20%20%20%20%20%20%20%20polymer_entity_count_nucleic_acid_hybrid%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_entry_group_membership%20%7B%0A%20%20%20%20%20%20%20%20group_id%0A%20%20%20%20%20%20%20%20aggregation_method%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20rcsb_binding_affinity%20%7B%0A%20%20%20%20%20%20%20%20comp_id%0A%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20value%0A%20%20%20%20%20%20%20%20unit%0A%20%20%20%20%20%20%20%20reference_sequence_identity%0A%20%20%20%20%20%20%20%20provenance_code%0A%20%20%20%20%20%20%20%20link%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20branched_entities%20%7B%0A%20%20%20%20%20%20%20%20rcsb_id%0A%20%20%20%20%20%20%20%20rcsb_branched_entity_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20%20%20entity_id%0A%20%20%20%20%20%20%20%20%20%20asym_ids%0A%20%20%20%20%20%20%20%20%20%20auth_asym_ids%0A%20%20%20%20%20%20%20%20%20%20reference_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20provenance_source%0A%20%20%20%20%20%20%20%20%20%20%20%20resource_name%0A%20%20%20%20%20%20%20%20%20%20%20%20resource_accession%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20prd%7B%0A%20%20%20%20%20%20%20%20%20%20rcsb_id%0A%20%20%20%20%20%20%20%20%20%20pdbx_reference_molecule%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20prd_id%0A%20%20%20%20%20%20%20%20%20%20%20%20chem_comp_id%0A%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20class%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20rcsb_branched_entity%20%7B%0A%20%20%20%20%20%20%20%20%20%20pdbx_description%0A%20%20%20%20%20%20%20%20%20%20formula_weight%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20pdbx_entity_branch%20%7B%0A%20%20%20%20%20%20%20%20%20%20rcsb_branched_component_count%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20branched_entity_instances%20%7B%0A%20%20%20%20%20%20%20%20%20%20rcsb_branched_entity_instance_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20%20%20%20%20entity_id%0A%20%20%20%20%20%20%20%20%20%20%20%20asym_id%0A%20%20%20%20%20%20%20%20%20%20%20%20auth_asym_id%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_branched_struct_conn%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20ordinal_id%0A%20%20%20%20%20%20%20%20%20%20%20%20role%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_branched_instance_feature%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20feature_value%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20comp_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20details%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20polymer_entities%20%7B%0A%20%20%20%20%20%20%20%20%20%20polymer_entity_instances%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_polymer_entity_instance_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20auth_asym_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20asym_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20entity_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_polymer_entity_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20%20%20%20%20entity_id%0A%20%20%20%20%20%20%20%20%20%20%20%20asym_ids%0A%20%20%20%20%20%20%20%20%20%20%20%20auth_asym_ids%0A%20%20%20%20%20%20%20%20%20%20%20%20uniprot_ids%0A%20%20%20%20%20%20%20%20%20%20%20%20reference_sequence_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20database_accession%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20uniprots%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_id%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_uniprot_protein%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20source_organism%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20scientific_name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_uniprot_external_reference%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20reference_name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20reference_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_polymer_entity%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20pdbx_description%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_ec_lineage%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20pdbx_ec%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_enzyme_class_combined%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20ec%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20provenance_source%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_polymer_entity_annotation%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20annotation_lineage%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20depth%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_polymer_entity_group_membership%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20group_id%0A%20%20%20%20%20%20%20%20%20%20%20%20similarity_cutoff%0A%20%20%20%20%20%20%20%20%20%20%20%20aggregation_method%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20entity_poly%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_entity_polymer_type%0A%20%20%20%20%20%20%20%20%20%20%20%20pdbx_seq_one_letter_code_can%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_sample_sequence_length%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_mutation_count%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_entity_source_organism%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20scientific_name%0A%20%20%20%20%20%20%20%20%20%20%20%20ncbi_scientific_name%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_gene_name%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20value%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20provenance_source%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_entity_host_organism%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20ncbi_scientific_name%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20prd%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_id%0A%20%20%20%20%20%20%20%20%20%20%20%20pdbx_reference_molecule%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20prd_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20chem_comp_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20class%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20chem_comp_nstd_monomers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20chem_comp%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20formula%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20mon_nstd_parent_comp_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20nonpolymer_entities%20%7B%0A%20%20%20%20%20%20%20%20%20%20rcsb_nonpolymer_entity_container_identifiers%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20%20%20%20%20entity_id%0A%20%20%20%20%20%20%20%20%20%20%20%20auth_asym_ids%0A%20%20%20%20%20%20%20%20%20%20%20%20asym_ids%0A%20%20%20%20%20%20%20%20%20%20%20%20nonpolymer_comp_id%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_nonpolymer_entity_annotation%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20nonpolymer_entity_instances%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_nonpolymer_entity_instance_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20auth_seq_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20auth_asym_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20asym_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20entry_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20entity_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20rcsb_nonpolymer_instance_validation_score%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20ranking_model_fit%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20ranking_model_geometry%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20average_occupancy%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20is_subject_of_investigation%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20is_subject_of_investigation_provenance%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20rcsb_nonpolymer_entity%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20pdbx_description%0A%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20nonpolymer_comp%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20chem_comp%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20formula_weight%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20formula%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20pdbx_reference_molecule%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20prd_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20chem_comp_id%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20name%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20class%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20rcsb_chem_comp_descriptor%20%7B%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20%20InChIKey%0A%20%20%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20assemblies%20%7B%0A%20%20%20%20%20%20%20%20rcsb_assembly_container_identifiers%20%7B%0A%20%20%20%20%20%20%20%20%20%20assembly_id%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20pdbx_struct_assembly%20%7B%0A%20%20%20%20%20%20%20%20%20%20rcsb_details%0A%20%20%20%20%20%20%20%20%20%20method_details%0A%20%20%20%20%20%20%20%20%20%20rcsb_candidate_assembly%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20pdbx_struct_assembly_auth_evidence%20%7B%0A%20%20%20%20%20%20%20%20%20%20experimental_support%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20rcsb_struct_symmetry%20%7B%0A%20%20%20%20%20%20%20%20%20%20kind%0A%20%20%20%20%20%20%20%20%20%20type%0A%20%20%20%20%20%20%20%20%20%20symbol%0A%20%20%20%20%20%20%20%20%20%20oligomeric_state%0A%20%20%20%20%20%20%20%20%20%20stoichiometry%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%20%20rcsb_assembly_info%20%7B%0A%20%20%20%20%20%20%20%20%20%20modeled_polymer_monomer_count%0A%20%20%20%20%20%20%20%20%7D%0A%20%20%20%20%20%20%7D%0A%20%20%7D%0A%7D%0A&variables=%7B%22id%22%3A%20%221YCR%22%7D"><button class="btn btn-sm btn-default"><span class="glyphicon glyphicon-cog"></span> Data API </button></a></div></div><h1><div class="results-content-type experimental"><span class="fa fa-flask" data-toggle="tooltip" data-placement="top" data-original-title="Experimental PDB Structure"></span></div><span id="structureID"> 1YCR</span></h1><h4><span id="structureTitle">MDM2 BOUND TO THE TRANSACTIVATION DOMAIN OF P53</span></h4><ul class="list-inline"><li id="header_doi"><strong>PDB DOI: </strong><a href="https://doi.org/10.2210/pdb1YCR/pdb" target="_blank" rel="noopener"> https://doi.org/10.2210/pdb1YCR/pdb</a></li></ul><ul class="list-unstyled"><li id="header_classification"><strong>Classification: <a href="/search?q=struct_keywords.pdbx_keywords:COMPLEX (ONCOGENE PROTEIN/PEPTIDE)">COMPLEX (ONCOGENE PROTEIN/PEPTIDE)</a></strong></li><li id="header_organism"><strong>Organism(s): </strong><a href="/search?q=rcsb_entity_source_organism.taxonomy_lineage.name:Homo sapiens">Homo sapiens</a></li><li id="header_expression-system"><strong>Expression System: </strong><a href="/search?q=rcsb_entity_host_organism.ncbi_scientific_name:Escherichia coli">Escherichia coli</a></li><!-- sum of mutations of all entities--><li id="header_mutation"><strong>Mutation(s): </strong>No <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="right" data-original-title="Indicates sequence mutations."></span></li><br><li id="header_deposited-released-dates"><strong>Deposited: </strong>1996-09-30 <strong>Released: </strong>1997-11-19 </li><li id="header_deposition-authors"><strong>Deposition Author(s): </strong><a href="/search?q=audit_author.name:Kussie, P.H.">Kussie, P.H.</a>, <a href="/search?q=audit_author.name:Pavletich, N.P.">Pavletich, N.P.</a></li></ul><hr><div class="row"><div class="col-sm-5 col-xs-12" id="header_experimentalDataSnapshot"><p><strong>Experimental Data Snapshot</strong></p><ul class="list-unstyled" id="exp_header_0_snapshot"><li id="exp_header_0_method"><strong>Method: </strong>X-RAY DIFFRACTION</li><li id="exp_header_0_diffraction_resolution"><strong>Resolution: </strong>2.60 Å</li><li id="exp_header_0_diffraction_rvalueFree"><strong>R-Value Free: </strong>0.276 </li><li id="exp_header_0_diffraction_rvalueWork"><strong>R-Value Work: </strong>0.200 </li><li id="exp_header_0_diffraction_rvalueObserved"><strong>R-Value Observed: </strong>0.200  </li></ul></div><div class="col-sm-7 col-xs-12"><!-- Experimental Validation HTML / PDF--><!-- Experimental Validation HTML / PDF--><style>div.validation-slider { border: 1px solid #ddd; padding: 5px; }
div.validation-slider img { max-width: 100%; }
</style><p><strong>wwPDB Validation</strong>  <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="right" data-original-title="For each metric, two values are noted, one for the percentile rank of the entry compared to the entire PDB archive and one compared with entries determined with the same experimental method. The blue side of the scale is considered "better" than those on the "worse" red side."> </span><span class="pull-right"><a class="btn btn-default btn-xs" href="/3d-view/1YCR?preset=validationReport"><i class="fa fa-cube"> </i>3D Report</a> <a class="btn btn-default btn-xs" target="_blank" rel="noopener" href="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_full_validation.pdf">Full Report</a></span></p><div class="validation-slider"><a target="_blank" rel="noopener" href="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_full_validation.pdf"><img src="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_multipercentile_validation.png" width="500"></a></div></div></div><br><div class="row"><div class="col-lg-12 col-md-12 col-sm-12 col-xs-12" id="currentVersionSection"><div class="well well-sm well-nomar">This is version 1.4 of the entry. See complete <a href="/versions/1YCR">history</a>. </div><br></div></div><br><div class="row"><div class="col-lg-12 col-md-12 col-sm-12 col-xs-12"><div class="panel panel-default" id="literaturepanel"><div class="panel-heading"><div class="panel-title">Literature<div class="btn-group pull-right"><button class="btn btn-default btn-xs hidden-print dropdown-toggle" type="button" data-toggle="dropdown" aria-expanded="false">Download Primary Citation <span class="caret"></span></button><ul class="dropdown-menu DropdownChangeGallery" role="menu"><li><a href="javascript:document.getElementsByTagName('body')[0].appendChild(document.createElement('script')).setAttribute('src','https://www.mendeley.com/minified/bookmarklet.js');"><img src="//cdn.rcsb.org/rcsb-pdb/explorer/SSPv2/images/MendeleyIcon.png" width="16"> Download Mendeley</a></li></ul></div></div></div><div class="panel-body"><div id="primarycitation"><h4>Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain.</h4><a class="querySearchLink" href="/search?q=citation.rcsb_authors:Kussie, P.H.">Kussie, P.H.</a>, <a class="querySearchLink" href="/search?q=citation.rcsb_authors:Gorina, S.">Gorina, S.</a>, <a class="querySearchLink" href="/search?q=citation.rcsb_authors:Marechal, V.">Marechal, V.</a>, <a class="querySearchLink" href="/search?q=citation.rcsb_authors:Elenbaas, B.">Elenbaas, B.</a>, <a class="querySearchLink" href="/search?q=citation.rcsb_authors:Moreau, J.">Moreau, J.</a>, <a class="querySearchLink" href="/search?q=citation.rcsb_authors:Levine, A.J.">Levine, A.J.</a>, <a class="querySearchLink" href="/search?q=citation.rcsb_authors:Pavletich, N.P.">Pavletich, N.P.</a><br><!-- if journal_abbrev already published--><p>(1996) Science <strong>274</strong>: 948-953</p><ul class="list-unstyled"><li id="pubmedLinks"><strong>PubMed</strong>: <a class="querySearchLink" href="/search?q=rcsb_pubmed_container_identifiers.pubmed_id:8875929">8875929</a> <span class="label label-external hidden-print"><a href="http://www.ncbi.nlm.nih.gov/pubmed/?term=8875929" target="_blank" rel="noopener">Search on PubMed</a></span></li><li id="pubmedDOI"><strong>DOI: </strong><a href="https://doi.org/10.1126/science.274.5289.948" target="_blank" rel="noopener">https://doi.org/10.1126/science.274.5289.948</a></li><li id="citationPrimaryRelatedStructures" style="word-wrap:break-word;">Primary Citation of Related Structures:  <br><a href="/structure/1YCQ">1YCQ</a>, <a href="/structure/1YCR">1YCR</a></li><!--TODO: add "Secondary Citation of Related Structures" when data is available --><br><li id="pubmedAbstractText"><strong>PubMed Abstract: </strong><br><div class="abstract" id="abstract"><p>The MDM2 oncoprotein is a cellular inhibitor of the p53 tumor suppressor in that it can bind the transactivation domain of p53 and downregulate its ability to activate transcription. In certain cancers, MDM2 amplification is a common event and contributes to the inactivation of p53. The crystal structure of the 109-residue amino-terminal domain of MDM2 bound to a 15-residue transactivation domain peptide of p53 revealed that MDM2 has a deep hydrophobic cleft on which the p53 peptide binds as an amphipathic alpha helix. The interface relies on the steric complementarity between the MDM2 cleft and the hydrophobic face of the p53 alpha helix and, in particular, on a triad of p53 amino acids-Phe19, Trp23, and Leu26-which insert deep into the MDM2 cleft. These same p53 residues are also involved in transactivation, supporting the hypothesis that MDM2 inactivates p53 by concealing its transactivation domain. The structure also suggests that the amphipathic alpha helix may be a common structural motif in the binding of a diverse family of transactivation factors to the TATA-binding protein-associated factors.</p></div><button class="btn btn-default btn-xs" onclick="toggleAbstract()" id="toggleBtn" style="margin-top: 3px"><span class="glyphicon glyphicon-plus-sign"></span> View More</button></li><hr><strong>Organizational Affiliation</strong>: <p>Cellular Biochemistry and Biophysics Program, Memorial Sloan-Kettering Cancer Center, New York, NY 10021, USA. nikola@xray2.mskcc.org</p></ul><!--journal_abbrev to be published--></div></div></div></div></div></div><div class="col-md-4 col-sm-12 col-xs-12"><script>// Used for Transmembrane Image Carousel
var structureFirstLigand = false;
var finalImageCount = 1;
var isNMR = false;
var modelCount = 1
var assemblies = ["1"]</script><div class="panel panel-default" id="galleryimagepanel"><div class="panel-body" id="structureimagesection"><div class="carousel slide" id="carousel-structuregallery" data-ride="carousel" data-interval="false"><div class="carousel-inner" role="listbox"><div class="item imageCarouselItem" id="Carousel-BiologicalUnit0"><div class="carousel-header">Asymmetric Unit</div><div class="clearfix"></div><img class="img-responsive center-block mainImage" src="data:image/gif;base64,R0lGODlhAQABAAD/ACwAAAAAAQABAAACADs=" style="image-rendering: -webkit-optimize-contrast;"><a class="btn btn-default btn-xs btn-enlargeImage" type="button" data-toggle="lightbox" data-gallery="multiimages0" href="#" data-title=""><span class="glyphicon glyphicon-resize-full" aria-hidden="true"></span></a><div class="galleryNewImages" id="AssemblyNewImage0"></div><div class="clearfix"></div><div class="carousel-footer"><p><assemblyid class="fa fa-cube"> </assemblyid><strong>3D View</strong>: <a href="/3d-view/1YCR/0">Structure</a> | <a href="/3d-sequence/1YCR?assemblyId=0">1D-3D View</a> | <span class="propernoun"><a href="/3d-view/1YCR?preset=validationReport">Validation Report</a></span></p></div></div><div class="item imageCarouselItem" id="Carousel-BiologicalUnit1"><div class="carousel-header">Biological Assembly 1  <a class="hidden-xs hidden-sm hidden-md icon-link" href="/docs/general-help/computed-structure-models-and-rcsborg#models-and-assembly"><span class="glyphicon glyphicon-question-sign" aria-hidden="true"></span></a></div><div class="clearfix"></div><img class="img-responsive center-block mainImage" src="data:image/gif;base64,R0lGODlhAQABAAD/ACwAAAAAAQABAAACADs=" style="image-rendering: -webkit-optimize-contrast;"><a class="btn btn-default btn-xs btn-enlargeImage" type="button" data-toggle="lightbox" data-gallery="multiimages1" href="#" data-title=""><span class="glyphicon glyphicon-resize-full" aria-hidden="true"></span></a><div class="galleryNewImages" id="AssemblyNewImage1"></div><div class="clearfix"></div><div class="carousel-footer"><p><assemblyid class="fa fa-cube"> </assemblyid><strong>3D View</strong>: <a href="/3d-view/1YCR/1">Structure</a> | <a href="/3d-sequence/1YCR?assemblyId=1">1D-3D View</a> | <span class="propernoun"><a href="/3d-view/1YCR?preset=validationReport">Validation Report</a></span></p><hr><strong>Global Symmetry</strong>: Asymmetric - C1 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Point group symmetry for the biological assembly. Chains are considered equivalent when their sequence identities are above 95%."></span><br><strong>Global Stoichiometry</strong>: Hetero 2-mer - <span style="word-wrap:break-word;">A1B1 </span><span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Chains with length below 5 residues or relatively short with respect to other chains are excluded from the stoichiometry and symmetry calculation."></span><div class="hidden-print" id="symmetryPart"></div><div class="hidden-print hide" id="symmetryFull"><br><button class="btn btn-default btn-xs hidden-print" id="full_symmetry_hide"><span class="glyphicon glyphicon-minus-sign"></span> Less</button></div><hr><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22structure%22%2C%22parameters%22%3A%7B%22value%22%3A%7B%22entry_id%22%3A%221YCR%22%2C%22assembly_id%22%3A%221%22%7D%7D%7D&return_type=assembly">Find Similar Assemblies</a><hr><p>Biological assembly 1 assigned by authors and generated by PISA (software)</p></div></div><a class="left carousel-control" href="#carousel-structuregallery" role="button" data-slide="prev"><span class="glyphicon glyphicon-chevron-left" aria-hidden="true"></span><span class="sr-only">Previous</span></a><a class="right carousel-control" href="#carousel-structuregallery" role="button" data-slide="next"><span class="glyphicon glyphicon-chevron-right" aria-hidden="true"></span><span class="sr-only">Next</span></a></div></div></div></div><!-- Show up only when there is a mix of structure type--><div class="well well-sm hidden-sm hidden-xs well-nomar" id="macromoleculeContent"><p><strong>Macromolecule Content</strong></p><ul><li id="contentStructureWeight">Total Structure Weight: 14.34 kDa <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Molecular weight (in kDa) of all non-water atoms in the deposited model, based on the full deposited sample sequence. Hydrogen atoms are included for the charged state in ARG, HIS & LYS residues."></span></li><li id="contentAtomSiteCount">Atom Count: 818 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Number of modeled non-hydrogen atoms in the deposited model."></span></li><li id="contentResidueCount">Modelled Residue Count: 98 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Number of modeled polymer monomers in the deposited model."></span></li><li id="contentResidueCount">Deposited Residue Count: 124 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Number of all (modeled or unmodeled) polymer monomers in the deposited model."></span></li><li id="contentProteinChainCount">Unique protein chains: 2</li></ul></div></div></div><br></div><div class="panel panel-default" id="macromoleculespanel"><div class="panel-heading"><div class="panel-title">Macromolecules </div></div><div class="panel-body"><div class="row"><div class="col-md-12 col-sm-12 col-xs-12"><div id="MacromoleculeTable"><div style="display: flow-root; margin-bottom: 5px;">Find similar proteins by: <div class="btn-group" role="group"><button class="btn-group btn btn-sm btn-default dropdown-toggle" id="dropdownCutOff" type="button" data-toggle="dropdown" aria-expanded="false" role="group" style="color: #337ab7; text-decoration: none;">Sequence <span class="caret"></span></button><ul class="dropdown-menu" role="menu" aria-labelledby="dropdownCutOff"><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A1%7D%7D&return_type=polymer_entity">100%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.95%7D%7D&return_type=polymer_entity">95%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.9%7D%7D&return_type=polymer_entity">90%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.8%7D%7D&return_type=polymer_entity">80%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.7%7D%7D&return_type=polymer_entity">70%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.6%7D%7D&return_type=polymer_entity">60%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.5%7D%7D&return_type=polymer_entity">50%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.4%7D%7D&return_type=polymer_entity">40%</a></li><li><a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22sequence%22%2C%22parameters%22%3A%7B%22target%22%3A%22pdb_protein_sequence%22%2C%22value%22%3A%22SQIPASEQETLVRPKPLLLKLLKSVGAQKDTYTMKEVLFYLGQYIMTKRLYDEKQQHIVYCSNDLLGDLFGVPSFSVKEHRKIYTMIYRNLVVVNQQESSDSGTSVSEN%22%2C%22identity_cutoff%22%3A0.3%7D%7D&return_type=polymer_entity">30%</a></li></ul></div> (by identity cutoff) | <a href="/search?query=%7B%22type%22%3A%22terminal%22%2C%22service%22%3A%22structure%22%2C%22parameters%22%3A%7B%22value%22%3A%7B%22entry_id%22%3A%221YCR%22%2C%22asym_id%22%3A%22A%22%7D%7D%7D&return_type=polymer_entity">3D Structure</a></div><div class="table-responsive"><a class="entity-anchor" name="entity-1"></a><table class="table table-bordered table-condensed tableEntity" id="table_macromolecule-protein-entityId-1"><tbody><tr class="info" style="color: #315880"><th colspan="6" style="padding-bottom: 0px; padding-top: 0px"><h5 style="font-weight: bold; font-size: 15px">Entity ID: 1</h5></th></tr><tr><th class="col-sm-2 col-lg-2 pad-by-five">Molecule</th><th class="col-sm-2 col-lg-2 pad-by-five">Chains<span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="If the two PDB chain IDs (label_asym_id; assigned by the PDB) and auth_asym_id (selected by the author) do not coincide, the chain ID is displayed as “label_asym_id [auth auth_asym_id]”"></span></th><th class="col-sm-2 col-lg-2 pad-by-five">Sequence Length</th><th class="col-sm-2 col-lg-2 pad-by-five">Organism</th><th class="col-sm-2 col-lg-2 pad-by-five">Details</th><th class="col-sm-2 col-lg-2 pad-by-five">Image</th></tr><tr id="macromolecule-entityId-1-rowDescription"><td>MDM2</td><td style="width:200px;"><div><a href="/sequence/1YCR#A">A</a></div></td><td>109</td><td><!-- Check if there is any data inside organism array--><a class="querySearchLink" href="/search?q=rcsb_entity_source_organism.taxonomy_lineage.name:Homo sapiens">Homo sapiens</a></td><td style="word-break: break-all;"><strong>Mutation(s)</strong>: 0 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="The number of mutations in this polymer sequence that are engineered from the reference sequence."></span><br><strong>Gene Names: </strong><a class="querySearchLink" href="/search?q=rcsb_entity_source_organism.rcsb_gene_name.value:MDM2">MDM2</a><br><strong>EC: </strong><a class="querySearchLink" href="/search?q=rcsb_polymer_entity.rcsb_ec_lineage.id:2.3.2.27">2.3.2.27</a><br></td><td style="text-align: center; vertical-align: middle"><a data-toggle="lightbox" data-title="1YCR_1: Represented by Chain A" href="https://cdn.rcsb.org/images/structures/1ycr_chain-A.jpeg"><img src="https://cdn.rcsb.org/images/structures/1ycr_chain-A.jpeg" style="width:100px;"></a></td></tr><tr><td colspan="6" style="padding-bottom: 0px; padding-top: 0px; background-color: #f5f5f5;"><h5 style="font-weight: bold; font-size: 15px">UniProt & NIH Common Fund Data Resources</h5></td></tr><tr><td colspan="6"><div class="col-lg-6 col-md-6 text-left" style="padding-left: 0">Find proteins for <a class="querySearchLink" href="/search?q=rcsb_polymer_entity_container_identifiers.reference_sequence_identifiers.database_accession:Q00987 AND rcsb_polymer_entity_container_identifiers.reference_sequence_identifiers.database_name:UniProt">Q00987</a> <i>(Homo sapiens)</i></div><div class="col-lg-3 col-md-3">Explore <span class="label label-rcsb"><a href="/groups/sequence/polymer_entity/Q00987">Q00987</a></span> <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Explore entries with the same UniProt reference sequence."></span></div><div class="col-lg-3 col-md-3 text-right" style="padding-right: 0">Go to UniProtKB:  <span class="label label-external"><a href="https://www.uniprot.org/uniprot/Q00987" target="_blank" rel="noopener">Q00987</a></span></div></td></tr><tr><td colspan="6"><div class="col-lg-4 col-md-4 text-left" style="padding-left: 0" title="Diseases, drugs and related data"><span>PHAROS:  <span class="label label-external"><a href="https://pharos.nih.gov/targets/Q00987" target="_blank" rel="noopener" onClick="sendGtag("SSP", "NIH", "PHAROS")">Q00987</a></span><break></break></span></div><div class="col-lg-4 col-md-4"></div><div class="col-lg-4 col-md-4 text-right" style="padding-right: 0"></div></td></tr><tr><td colspan="6" style="padding-bottom: 0px; padding-top: 0px; background-color: #f5f5f5;"><h5 style="font-weight: bold; font-size: 15px">Entity Groups
 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="This entity has been grouped together with similar entities from other structures. Use the links below to navigate to the corresponding group pages and explore the characteristics of similar entities." style="vertical-align: bottom;"></span></h5></td></tr><tr><td class="col-lg-2 col-md-2">Sequence Clusters</td><td class="col-lg-10 col-md-10 group-membership" colspan="5"><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/1047_30">30% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/902_50">50% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/651_70">70% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/460_90">90% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/426_95">95% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/21178_100">100% Identity<span class="fa fa-clone"></span></a></span></td></tr><tr><td class="col-lg-2 col-md-2">UniProt Group</td><td class="col-lg-10 col-md-10 group-membership" colspan="5"><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/Q00987">Q00987<span class="fa fa-clone"></span></a></span></td></tr><tr><td colspan="6" style="padding-bottom: 0px; padding-top: 0px; background-color: #f5f5f5;"><h5 style="font-weight: bold; font-size: 15px; display:inline-block;float:left">Protein Feature View</h5><h5 style="font-weight: bold; font-size: 15px; display:inline-block;float:right;"><a href="/sequence/1YCR#A">Expand</a></h5></td></tr><tr style="border-bottom: 5px solid #ddd" id="RcsbFv"><td class="ProteinFeatureView" colspan="6"><div id="entity" style="margin-top:20px"><div class="col-xs-12 col-sm-12 col-md-4 col-lg-4 form-group" style="margin-bottom: 0; padding-left: 0"><ul class="list-group" style="margin-bottom:0;display:inline-block;width:50%;"><li class="list-group-item" style="border-color:#fff;"><strong>Reference Sequence</strong></li></ul><div id="rcsbWebAppSelect_1" style="display:inline-block;"></div><div class="material-switch pull-right"></div></div></div><div class="clearfix"></div><div id="rcsbWebApp_1" style="margin-bottom: 50px;"></div><script>var headers = {
"Rcsb-Analytics-Traffic-Origin": "internal",
"Rcsb-Analytics-Traffic-Stage": "saguaro"
};
RcsbFvWebApp.RcsbRequestContextManager.initializeBorregoClient({
requestConfig: {
headers
}
});
RcsbFvWebApp.RcsbRequestContextManager.initializeYosemiteClient({
requestConfig: {
headers
}
});
RcsbFvWebApp.RcsbRequestContextManager.initializeArchesClient({
requestConfig: {
headers
}
});</script><script>setTimeout(function(){
RcsbFvWebApp.buildEntitySummaryFv(
"rcsbWebApp_1",
"rcsbWebAppSelect_1",
"1YCR_1"
);
},100)
</script></td></tr></tbody></table></div><p class="text-muted">Find similar proteins by: Sequence | 3D Structure <span class="fa fa-info-circle" aria-hidden="true" data-toggle="tooltip" data-placement="top" title="Sequence and structure search is not available for sequences with length less than or equal to 25"></span></p><div class="table-responsive"><a class="entity-anchor" name="entity-2"></a><table class="table table-bordered table-condensed tableEntity" id="table_macromolecule-protein-entityId-2"><tbody><tr class="info" style="color: #315880"><th colspan="6" style="padding-bottom: 0px; padding-top: 0px"><h5 style="font-weight: bold; font-size: 15px">Entity ID: 2</h5></th></tr><tr><th class="col-sm-2 col-lg-2 pad-by-five">Molecule</th><th class="col-sm-2 col-lg-2 pad-by-five">Chains<span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="If the two PDB chain IDs (label_asym_id; assigned by the PDB) and auth_asym_id (selected by the author) do not coincide, the chain ID is displayed as “label_asym_id [auth auth_asym_id]”"></span></th><th class="col-sm-2 col-lg-2 pad-by-five">Sequence Length</th><th class="col-sm-2 col-lg-2 pad-by-five">Organism</th><th class="col-sm-2 col-lg-2 pad-by-five">Details</th><th class="col-sm-2 col-lg-2 pad-by-five">Image</th></tr><tr id="macromolecule-entityId-2-rowDescription"><td>P53</td><td style="width:200px;"><div><a href="/sequence/1YCR#B">B</a></div></td><td>15</td><td><!-- Check if there is any data inside organism array--><small class="text-muted"><i>N/A</i></small></td><td style="word-break: break-all;"><strong>Mutation(s)</strong>: 0 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="The number of mutations in this polymer sequence that are engineered from the reference sequence."></span><br></td><td style="text-align: center; vertical-align: middle"><a data-toggle="lightbox" data-title="1YCR_2: Represented by Chain B" href="https://cdn.rcsb.org/images/structures/1ycr_chain-B.jpeg"><img src="https://cdn.rcsb.org/images/structures/1ycr_chain-B.jpeg" style="width:100px;"></a></td></tr><tr><td colspan="6" style="padding-bottom: 0px; padding-top: 0px; background-color: #f5f5f5;"><h5 style="font-weight: bold; font-size: 15px">UniProt & NIH Common Fund Data Resources</h5></td></tr><tr><td colspan="6"><div class="col-lg-6 col-md-6 text-left" style="padding-left: 0">Find proteins for <a class="querySearchLink" href="/search?q=rcsb_polymer_entity_container_identifiers.reference_sequence_identifiers.database_accession:P04637 AND rcsb_polymer_entity_container_identifiers.reference_sequence_identifiers.database_name:UniProt">P04637</a> <i>(Homo sapiens)</i></div><div class="col-lg-3 col-md-3">Explore <span class="label label-rcsb"><a href="/groups/sequence/polymer_entity/P04637">P04637</a></span> <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="Explore entries with the same UniProt reference sequence."></span></div><div class="col-lg-3 col-md-3 text-right" style="padding-right: 0">Go to UniProtKB:  <span class="label label-external"><a href="https://www.uniprot.org/uniprot/P04637" target="_blank" rel="noopener">P04637</a></span></div></td></tr><tr><td colspan="6"><div class="col-lg-4 col-md-4 text-left" style="padding-left: 0" title="Diseases, drugs and related data"><span>PHAROS:  <span class="label label-external"><a href="https://pharos.nih.gov/targets/P04637" target="_blank" rel="noopener" onClick="sendGtag("SSP", "NIH", "PHAROS")">P04637</a></span><break></break></span></div><div class="col-lg-4 col-md-4"></div><div class="col-lg-4 col-md-4 text-right" style="padding-right: 0"></div></td></tr><tr><td colspan="6" style="padding-bottom: 0px; padding-top: 0px; background-color: #f5f5f5;"><h5 style="font-weight: bold; font-size: 15px">Entity Groups
 <span class="glyphicon glyphicon-info-sign hidden-xs hidden-sm hidden-md" aria-hidden="true" data-toggle="tooltip" data-placement="top" data-original-title="This entity has been grouped together with similar entities from other structures. Use the links below to navigate to the corresponding group pages and explore the characteristics of similar entities." style="vertical-align: bottom;"></span></h5></td></tr><tr><td class="col-lg-2 col-md-2">Sequence Clusters</td><td class="col-lg-10 col-md-10 group-membership" colspan="5"><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/19160_30">30% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/25164_50">50% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/24653_70">70% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/20146_90">90% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/18405_95">95% Identity<span class="fa fa-clone"></span></a></span><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/10399_100">100% Identity<span class="fa fa-clone"></span></a></span></td></tr><tr><td class="col-lg-2 col-md-2">UniProt Group</td><td class="col-lg-10 col-md-10 group-membership" colspan="5"><span class="label label-rcsb"><a href="/groups/summary/polymer_entity/P04637">P04637<span class="fa fa-clone"></span></a></span></td></tr><tr><td colspan="6" style="padding-bottom: 0px; padding-top: 0px; background-color: #f5f5f5;"><h5 style="font-weight: bold; font-size: 15px; display:inline-block;float:left">Protein Feature View</h5><h5 style="font-weight: bold; font-size: 15px; display:inline-block;float:right;"><a href="/sequence/1YCR#B">Expand</a></h5></td></tr><tr style="border-bottom: 5px solid #ddd" id="RcsbFv"><td class="ProteinFeatureView" colspan="6"><div id="entity" style="margin-top:20px"><div class="col-xs-12 col-sm-12 col-md-4 col-lg-4 form-group" style="margin-bottom: 0; padding-left: 0"><ul class="list-group" style="margin-bottom:0;display:inline-block;width:50%;"><li class="list-group-item" style="border-color:#fff;"><strong>Reference Sequence</strong></li></ul><div id="rcsbWebAppSelect_2" style="display:inline-block;"></div><div class="material-switch pull-right"></div></div></div><div class="clearfix"></div><div id="rcsbWebApp_2" style="margin-bottom: 50px;"></div><script>var headers = {
"Rcsb-Analytics-Traffic-Origin": "internal",
"Rcsb-Analytics-Traffic-Stage": "saguaro"
};
RcsbFvWebApp.RcsbRequestContextManager.initializeBorregoClient({
requestConfig: {
headers
}
});
RcsbFvWebApp.RcsbRequestContextManager.initializeYosemiteClient({
requestConfig: {
headers
}
});
RcsbFvWebApp.RcsbRequestContextManager.initializeArchesClient({
requestConfig: {
headers
}
});</script><script>setTimeout(function(){
RcsbFvWebApp.buildEntitySummaryFv(
"rcsbWebApp_2",
"rcsbWebAppSelect_2",
"1YCR_2"
);
},100)
</script></td></tr></tbody></table></div></div></div></div></div></div><div class="panel panel-default" id="experimentaldata-validation"><div class="panel-heading"><div class="panel-title">Experimental Data & Validation</div></div><div class="panel-body"><div class="row"><div class="col-md-6 col-sm-12 col-xs-12"><h4>Experimental Data</h4><div class="row"><ul class="list-unstyled col-sm-12 col-lg-12 col-md-12" id="experimentaldatabottom"><li id="exp_header_0_method"><strong>Method: </strong>X-RAY DIFFRACTION</li><li id="exp_header_0_diffraction_resolution"><strong>Resolution: </strong>2.60 Å</li><li id="exp_header_0_diffraction_rvalueFree"><strong>R-Value Free: </strong>0.276 </li><li id="exp_header_0_diffraction_rvalueWork"><strong>R-Value Work: </strong>0.200 </li><li id="exp_header_0_diffraction_rvalueObserved"><strong>R-Value Observed: </strong>0.200 </li><li id="exp_undefined_xray_spaceGroup"><strong>Space Group: <a class="querySearchLink" href="/search?q=symmetry.space_group_name_H_M:C 2 2 21">C 2 2 2<sub>1</sub></a></strong></li></ul><div class="col-lg-12 col-md-12 col-sm-12"><strong>Unit Cell</strong>:<table class="table table-bordered table-condensed" id="unitCellTable"><thead><tr class="active"><th>Length ( Å )</th><th>Angle ( ˚ )</th></tr></thead><tbody><tr><td>a = 43.414</td><td>α = 90</td></tr><tr><td>b = 100.546</td><td>β = 90</td></tr><tr><td>c = 54.853</td><td>γ = 90</td></tr></tbody></table></div></div><strong>Software Package:</strong><table class="table table-bordered table-condensed" id="xraysoftwarePackages"><thead><tr class="active"><th>Software Name</th><th>Purpose</th></tr></thead><tbody><tr><td>X-PLOR</td><td>refinement</td></tr><tr><td>TNT</td><td>refinement</td></tr><tr><td>X-PLOR</td><td>model building</td></tr><tr><td>HKL</td><td>data reduction</td></tr><tr><td>X-PLOR</td><td>phasing</td></tr></tbody></table></div><div class="col-md-6 col-sm-12 col-xs-12"><!-- Experimental Validation HTML / PDF--><h4>Structure Validation</h4><!-- NOTE: check pdx/idf/ftp.json file--><p id="validationSection">View <a target="_blank" rel="noopener" href="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_full_validation.pdf">Full Validation Report</a></p><a target="_blank" rel="noopener" href="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_full_validation.pdf"><img class="img-thumbnail" src="//files.rcsb.org/pub/pdb/validation_reports/yc/1ycr/1ycr_multipercentile_validation.png" width="500"></a><br></div></div><br><div class="row"><div class="col-md-6 col-sm-12 col-xs-12"><a class="btn btn-sm btn-default hidden-print" href="/experimental/1YCR">View more in-depth experimental data</a></div></div></div></div><div class="panel panel-default" id="entry-history"><div class="panel-heading"><div class="panel-title">Entry History </div></div><div class="panel-body"><div class="row"><div class="col-md-6 col-sm-6 col-xs-12 col-xs-12"><h4>Deposition Data</h4><ul class="list-unstyled" id="DepositionDataList"><li><strong>Released Date: </strong>1997-11-19 </li><strong>Deposition Author(s): </strong><a href="/search?q=audit_author.name:Kussie, P.H.">Kussie, P.H.</a>, <a href="/search?q=audit_author.name:Pavletich, N.P.">Pavletich, N.P.</a></ul></div><div class="col-md-6 col-sm-6 col-xs-12 col-xs-12"><h4>Revision History <small><a href="/versions/1YCR">(Full details and data files)</a></small></h4><ul id="revisionHistorySection"><li><strong>Version 1.0: 1997-11-19</strong><br>Type: Initial release<br></li><li><strong>Version 1.1: 2008-03-24</strong><br>Changes: Version format compliance </li><li><strong>Version 1.2: 2011-07-13</strong><br>Changes: Version format compliance </li><li><strong>Version 1.3: 2019-07-17</strong><br>Changes: Data collection, Other, Refinement description </li><li><strong>Version 1.4: 2019-08-14</strong><br>Changes: Data collection </li></ul></div></div></div></div></div><div class="clearfix"></div><script>$("#structuresummarytab").addClass("active");</script><script src="/js/bootstrap-slider.min.js"></script><script src="/js/jquery.svg.js"></script><script src="/js/structure/helper.js"></script><script src="/js/structure/image_carousel_helper.js"></script><script src="/js/structure/poseview.js"></script></div><div data-elastic-exclude><div class="hidden-print" id="footer_main"><div class="container"><div class="row" style="padding-left:0; margin-left:0"><div class="col-md-6 col-sm-12"><div class="row"> <div class="col-sm-4" style="padding-left:0"><ul><li><span class="row_header">About</span></li><li><a href="/pages/about-us/index">About Us</a></li><li><a href="/pages/policies#References">Citing Us</a></li><li><a href="/pages/publications">Publications</a></li><li><a href="/pages/team">Team</a></li><li><a href="/pages/jobs">Careers</a></li><li><a href="/pages/policies">Usage & Privacy</a></li></ul></div><div class="col-sm-4" style="padding-left:0"><ul><li><span class="row_header">Help</span></li><li><a href="/pages/contactus">Contact Us</a></li><li><a href="/docs/general-help/organization-of-3d-structures-in-the-protein-data-bank">Documentation</a></li><li><a href="/docs/general-help/website-faq">Website FAQ</a></li><li><a href="/docs/general-help/glossary">Glossary</a></li><li><a href="https://status.rcsb.org/" target="_blank">Service Status</a></li></ul></div><div class="col-md-4 hidden-sm"></div></div></div><div class="col-md-6 col-sm-12"><div class="row"> <div class="col-sm-4"><p class="row_header">RCSB PDB (<a href="pages/policies#References">citation</a>) is hosted by</p><div class="footerLogos"><hr><a href="http://www.rutgers.edu/"> <img class="Rutgerslogo" src="https://cdn.rcsb.org/rcsb-pdb//v2/common/images/Logo_Rutgers.png" alt="Rutgers University logo"></a><hr><a href="https://ucsd.edu/"> <img class="UCSDlogo" src="https://cdn.rcsb.org/rcsb-pdb//v2/common/images/Logo_UCSD-navy.png" alt="Uiversity of California San Diego logo"></a><a href="https://www.sdsc.edu/"> <img class="SDSClogo" src="https://cdn.rcsb.org/rcsb-pdb//v2/common/images/Logo_SDSC-black.png" alt="San Diego Supercomputer Center logo"></a><hr><a href="https://www.ucsf.edu/"> <img class="UCSFlogo" src="https://cdn.rcsb.org/rcsb-pdb//v2/common/images/Logo_UCSF.png" alt="University of California San Francisco Logo"></a></div></div><div class="col-sm-4"><p class="row_header">RCSB PDB is a member of the</p><div class="footerLogos"><span id="pdbmembers_footer"><hr><a href="https://www.wwpdb.org" target="_blank" rel="noopener"><img class="wwpdblogo" src="https://cdn.rcsb.org/rcsb-pdb//v2/common/images/Logo_wwpdb.png" alt="wwPDB"></a><hr><a href="https://www.emdataresource.org" target="_blank" rel="noopener"><img id="emdatabanklogo_footer" src="https://cdn.rcsb.org/rcsb-pdb//v2/common/images/EMDataResourcelogo.png" alt="EMDataResource"></a></span></div></div><div class="col-sm-4"><ul><li><span class="row_header">RCSB Partners</span></li><li><a href="https://nakb.org/" target="_blank" rel="noopener">Nucleic Acid Knowledgebase</a></li></ul><ul><li><span class="row_header"><a href="https://www.wwpdb.org/" target="_blank" rel="noopener" style="text-decoration: none; color: black">wwPDB Partners</a></span></li><li><a href="/">RCSB PDB</a></li><li><a href="https://www.ebi.ac.uk/pdbe/" target="_blank" rel="noopener">PDBe</a></li><li><a href="https://www.pdbj.org/" target="_blank" rel="noopener">PDBj</a></li><li><a href="https://bmrb.io/" target="_blank" rel="noopener">BMRB</a></li><li><a href="https://emdb-empiar.org/" target="_blank" rel="noopener">EMDB</a></li></ul></div></div></div></div></div></div><div id="footer_grant"><div class="container"><div class="row"><p>RCSB PDB Core Operations are funded by
the <a href="http://www.nsf.gov/" target="_blank" rel="noopener">National Science Foundation</a> (DBI-1832184),
the <a href="http://science.energy.gov/" target="_blank" rel="noopener">US Department of Energy</a> (DE-SC0019749),
and the <a href="http://www.cancer.gov/" target="_blank" rel="noopener">National Cancer Institute</a>,
<a href="http://www.niaid.nih.gov/" target="_blank" rel="noopener">National Institute of Allergy and Infectious Diseases</a>,
and <a href="http://www.nigms.nih.gov/" target="_blank" rel="noopener">National Institute of General Medical Sciences</a> of the <a href="http://www.nih.gov/" target="_blank" rel="noopener">National Institutes of Health</a> under grant R01GM133198.</p></div></div></div><div id="jira-fdbck"></div></div></div></body></html>
Encoding Data¶
URLs are only allowed to have alphanumeric characters (and a handful of punctuation marks). This means data needs to be encoded when passing it as a value. requests will do this for you if you pass values as a dictionary.
values = {'q':"What's the meaning of life?"}
data = requests.get('http://www.google.com/search',values)
data.url
'http://www.google.com/search?q=What%27s+the+meaning+of+life%3F'
import requests, re
page = requests.get('https://www.whatsmybrowser.org/').text
re.findall(r'You’re using (.*?)\.',page)
['python-requests 2', 'python-requests 2']
page = requests.get('https://www.whatsmybrowser.org/',headers={'User-Agent': 'Mozilla/5.0 (Macintosh; Intel Mac OS X 10_12_6) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/61.0.3163.100 Safari/537.36'}).text
re.findall(r'You’re using (.*?)\.',page)
['Chrome 61', 'Chrome 61']
Use mechanize if a site is giving you trouble
Getting Data From the Web¶
If it's on the web, you can get it into python.
- screen scraping - getting data from your computer screen; often to the extreme of using screen shots and OCR
- web scraping - downloading HTML content and parsing out what you need
Web Scraping¶
The advantage of web scraping is that it always works - if you can see the data in your browser, you can see it in python.
Disadvantages:
HTMLParser isn't very sophisticated and requires you to manage context
- see pyquery for a more powerful HTML parsing package inspired by JQuery
- BeautifulSoup is another popular library for extracting information from HTML
Scraping is slow and inefficient
- may need to make several requests to get your data (think pagination)
- downloads a lot more than just your data (html)
- your code may be easily broken by changes in the website design
Only parse raw HTML if there is no better option
ReSTful Web Services¶
REpresentational State Transfer
Client–server
Stateless
Cacheable
Layered system
Uniform interface
- basically, resources are identified by their url
- ReST does not specify the format of the resources, but they are often provided in XML
XML¶
Extensible Markup Language. A very general way to express structured data; a generalization of HTML.
Tag Key building block of XML - starts and ends with < >
<div>start tag</div>end tag<br />empty element tag (not matching end)
Element A component of the document; everything between a start and end tag. May contain child elements.
Attribute A name-value pair within a start or empty element tag
XML Example¶
Let's consider accessing NCBI's Entrez service to get data from the Gene Expression Omnibus (note we could use BioPython and avoid doing this directly).
Let's look for data from humans with between 100 and 500 samples:
- (human[Organism]) AND 100:500[Number of Samples]
We construct a URL to represent this query.
db=gds - GEO datasets
- GEO DataSets is a study-level database which users can search for studies relevant to their interests. The database stores descriptions of all original submitter-supplied records, as well as curated DataSets.
result = requests.get(\
"http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esearch.fcgi",\
params={'db':'gds',\
'term':'human[Organism] AND 100:500[Number of Samples]'}).text
print(result[:297])
<?xml version="1.0" encoding="UTF-8" ?> <!DOCTYPE eSearchResult PUBLIC "-//NLM//DTD esearch 20060628//EN" "https://eutils.ncbi.nlm.nih.gov/eutils/dtd/20060628/esearch.dtd"> <eSearchResult><Count>6651</Count><RetMax>20</RetMax><RetStart>0</RetStart><IdList> <Id>200245735</Id> <Id>200241425</Id> <I
%%html
<div id="xmlq" style="width: 500px"></div>
<script>
var divid = '#xmlq';
jQuery(divid).asker({
id: divid,
question: "On the previous slide, RetMax is what?",
answers: ['Element','Attribute','Tag','Component'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
xml.etree.ElementTree¶
import xml.etree.ElementTree as ET
root = ET.fromstring(result)
print(root.tag,root.attrib)
eSearchResult {}
for child in root:
print(child.tag)
Count RetMax RetStart IdList TranslationSet TranslationStack QueryTranslation
root.find('IdList')
<Element 'IdList' at 0x7f2918f418f0>
ids = [child.text for child in root.find('IdList')]
print(ids)
['200245735', '200241425', '200181711', '200181709', '200245630', '200241428', '200232216', '200231719', '200245626', '200240155', '200222110', '200185796', '200233715', '200227832', '200226400', '200211631', '200211158', '200131747', '200237999', '200225817']
root.findall('Id') #find only considers children
[]
ids = [elem.text for elem in root.iter('Id')] #iter considers the full tree
print(ids)
['200245735', '200241425', '200181711', '200181709', '200245630', '200241428', '200232216', '200231719', '200245626', '200240155', '200222110', '200185796', '200233715', '200227832', '200226400', '200211631', '200211158', '200131747', '200237999', '200225817']
XML Parsing Alternative: Regular Expressions¶
print(result[:297])
<?xml version="1.0" encoding="UTF-8" ?> <!DOCTYPE eSearchResult PUBLIC "-//NLM//DTD esearch 20060628//EN" "https://eutils.ncbi.nlm.nih.gov/eutils/dtd/20060628/esearch.dtd"> <eSearchResult><Count>6651</Count><RetMax>20</RetMax><RetStart>0</RetStart><IdList> <Id>200245735</Id> <Id>200241425</Id> <I
import re
regex = re.compile(r'<Id>(\d+)</Id>')
print(regex.search(result))
<re.Match object; span=(257, 275), match='<Id>200245735</Id>'>
ids = [m.group(1) for m in regex.finditer(result)]
print(ids)
['200245735', '200241425', '200181711', '200181709', '200245630', '200241428', '200232216', '200231719', '200245626', '200240155', '200222110', '200185796', '200233715', '200227832', '200226400', '200211631', '200211158', '200131747', '200237999', '200225817']
JSON: JavaScript Object Notation¶
A lightweight data-interchange format.
Essentially, represent data as JavaScript, which is very similar to python dictionaries/lists.
import json
json.dumps({'a':1, 'b':[1.2,1.3,1.4]})
'{"a": 1, "b": [1.2, 1.3, 1.4]}'
PDB Restful¶
https://data.rcsb.org/redoc/index.html
Access PDB data through endpoints (URLs). The path of the endpoints starts with https://data.rcsb.org/rest/v1/core, followed by the type of the resource, e.g. entry, polymer_entity, and the identifier.
response = requests.get('https://data.rcsb.org/rest/v1/core/entry/4hhb')
print(response.text)
{"audit_author":[{"name":"Fermi, G.","pdbx_ordinal":1},{"name":"Perutz, M.F.","pdbx_ordinal":2}],"cell":{"angle_alpha":90.0,"angle_beta":99.34,"angle_gamma":90.0,"length_a":63.15,"length_b":83.59,"length_c":53.8,"zpdb":4},"citation":[{"country":"UK","id":"primary","journal_abbrev":"J.Mol.Biol.","journal_id_astm":"JMOBAK","journal_id_csd":"0070","journal_id_issn":"0022-2836","journal_volume":"175","page_first":"159","page_last":"174","pdbx_database_id_doi":"10.1016/0022-2836(84)90472-8","pdbx_database_id_pub_med":6726807,"rcsb_authors":["Fermi, G.","Perutz, M.F.","Shaanan, B.","Fourme, R."],"rcsb_is_primary":"Y","rcsb_journal_abbrev":"J Mol Biol","title":"The crystal structure of human deoxyhaemoglobin at 1.74 A resolution","year":1984},{"country":"UK","id":"1","journal_abbrev":"Nature","journal_id_astm":"NATUAS","journal_id_csd":"0006","journal_id_issn":"0028-0836","journal_volume":"295","page_first":"535","rcsb_authors":["Perutz, M.F.","Hasnain, S.S.","Duke, P.J.","Sessler, J.L.","Hahn, J.E."],"rcsb_is_primary":"N","rcsb_journal_abbrev":"Nature","title":"Stereochemistry of Iron in Deoxyhaemoglobin","year":1982},{"country":"US","id":"3","journal_abbrev":"Annu.Rev.Biochem.","journal_id_astm":"ARBOAW","journal_id_csd":"0413","journal_id_issn":"0066-4154","journal_volume":"48","page_first":"327","rcsb_authors":["Perutz, M.F."],"rcsb_is_primary":"N","rcsb_journal_abbrev":"Annu Rev Biochem","title":"Regulation of Oxygen Affinity of Hemoglobin. Influence of Structure of the Globin on the Heme Iron","year":1979},{"country":"UK","id":"4","journal_abbrev":"J.Mol.Biol.","journal_id_astm":"JMOBAK","journal_id_csd":"0070","journal_id_issn":"0022-2836","journal_volume":"100","page_first":"3","rcsb_authors":["Teneyck, L.F.","Arnone, A."],"rcsb_is_primary":"N","rcsb_journal_abbrev":"J Mol Biol","title":"Three-Dimensional Fourier Synthesis of Human Deoxyhemoglobin at 2.5 Angstroms Resolution, I.X-Ray Analysis","year":1976},{"country":"UK","id":"5","journal_abbrev":"J.Mol.Biol.","journal_id_astm":"JMOBAK","journal_id_csd":"0070","journal_id_issn":"0022-2836","journal_volume":"97","page_first":"237","rcsb_authors":["Fermi, G."],"rcsb_is_primary":"N","rcsb_journal_abbrev":"J Mol Biol","title":"Three-Dimensional Fourier Synthesis of Human Deoxyhaemoglobin at 2.5 Angstroms Resolution, Refinement of the Atomic Model","year":1975},{"country":"UK","id":"6","journal_abbrev":"Nature","journal_id_astm":"NATUAS","journal_id_csd":"0006","journal_id_issn":"0028-0836","journal_volume":"228","page_first":"516","rcsb_authors":["Muirhead, H.","Greer, J."],"rcsb_is_primary":"N","rcsb_journal_abbrev":"Nature","title":"Three-Dimensional Fourier Synthesis of Human Deoxyhaemoglobin at 3.5 Angstroms Resolution","year":1970},{"book_publisher":"Oxford University Press","id":"2","journal_abbrev":"Haemoglobin and Myoglobin. Atlas of Molecular Structures in Biology","journal_id_csd":"0986","journal_id_issn":"0-19-854706-4","journal_volume":"2","rcsb_authors":["Fermi, G.","Perutz, M.F."],"rcsb_is_primary":"N","rcsb_journal_abbrev":"Haemoglobin And Myoglobin Atlas Of Molecular Structures In Biology","year":1981},{"book_publisher":"National Biomedical Research Foundation, Silver Spring,Md.","id":"7","journal_abbrev":"Atlas of Protein Sequence and Structure (Data Section)","journal_id_csd":"0435","journal_id_issn":"0-912466-02-2","journal_volume":"5","page_first":"56","rcsb_is_primary":"N","rcsb_journal_abbrev":"Atlas Of Protein Sequence And Structure (data Section)","year":1972},{"book_publisher":"National Biomedical Research Foundation, Silver Spring,Md.","id":"8","journal_abbrev":"Atlas of Protein Sequence and Structure (Data Section)","journal_id_csd":"0435","journal_id_issn":"0-912466-02-2","journal_volume":"5","page_first":"64","rcsb_is_primary":"N","rcsb_journal_abbrev":"Atlas Of Protein Sequence And Structure (data Section)","year":1972}],"diffrn":[{"crystal_id":"1","id":"1"}],"entry":{"id":"4HHB"},"exptl":[{"method":"X-RAY DIFFRACTION"}],"exptl_crystal":[{"density_matthews":2.26,"density_percent_sol":45.48,"id":"1"}],"pdbx_audit_revision_category":[{"category":"atom_site","data_content_type":"Structure model","ordinal":1,"revision_ordinal":4},{"category":"database_PDB_caveat","data_content_type":"Structure model","ordinal":2,"revision_ordinal":4},{"category":"entity","data_content_type":"Structure model","ordinal":3,"revision_ordinal":4},{"category":"entity_name_com","data_content_type":"Structure model","ordinal":4,"revision_ordinal":4},{"category":"entity_src_gen","data_content_type":"Structure model","ordinal":5,"revision_ordinal":4},{"category":"pdbx_database_status","data_content_type":"Structure model","ordinal":6,"revision_ordinal":4},{"category":"pdbx_validate_rmsd_angle","data_content_type":"Structure model","ordinal":7,"revision_ordinal":4},{"category":"pdbx_validate_rmsd_bond","data_content_type":"Structure model","ordinal":8,"revision_ordinal":4},{"category":"struct_ref","data_content_type":"Structure model","ordinal":9,"revision_ordinal":4},{"category":"struct_ref_seq","data_content_type":"Structure model","ordinal":10,"revision_ordinal":4},{"category":"atom_site","data_content_type":"Structure model","ordinal":11,"revision_ordinal":5},{"category":"pdbx_validate_rmsd_angle","data_content_type":"Structure model","ordinal":12,"revision_ordinal":5},{"category":"pdbx_validate_rmsd_bond","data_content_type":"Structure model","ordinal":13,"revision_ordinal":5},{"category":"struct_site","data_content_type":"Structure model","ordinal":14,"revision_ordinal":5},{"category":"atom_site","data_content_type":"Structure model","ordinal":15,"revision_ordinal":6},{"category":"atom_sites","data_content_type":"Structure model","ordinal":16,"revision_ordinal":6},{"category":"database_2","data_content_type":"Structure model","ordinal":17,"revision_ordinal":6},{"category":"database_PDB_matrix","data_content_type":"Structure model","ordinal":18,"revision_ordinal":6},{"category":"pdbx_struct_conn_angle","data_content_type":"Structure model","ordinal":19,"revision_ordinal":6},{"category":"pdbx_validate_close_contact","data_content_type":"Structure model","ordinal":20,"revision_ordinal":6},{"category":"pdbx_validate_main_chain_plane","data_content_type":"Structure model","ordinal":21,"revision_ordinal":6},{"category":"pdbx_validate_peptide_omega","data_content_type":"Structure model","ordinal":22,"revision_ordinal":6},{"category":"pdbx_validate_planes","data_content_type":"Structure model","ordinal":23,"revision_ordinal":6},{"category":"pdbx_validate_polymer_linkage","data_content_type":"Structure model","ordinal":24,"revision_ordinal":6},{"category":"pdbx_validate_rmsd_angle","data_content_type":"Structure model","ordinal":25,"revision_ordinal":6},{"category":"pdbx_validate_rmsd_bond","data_content_type":"Structure model","ordinal":26,"revision_ordinal":6},{"category":"pdbx_validate_torsion","data_content_type":"Structure model","ordinal":27,"revision_ordinal":6},{"category":"struct_ncs_oper","data_content_type":"Structure model","ordinal":28,"revision_ordinal":6},{"category":"pdbx_database_remark","data_content_type":"Structure model","ordinal":29,"revision_ordinal":7}],"pdbx_audit_revision_details":[{"data_content_type":"Structure model","ordinal":1,"provider":"repository","revision_ordinal":1,"type":"Initial release"},{"data_content_type":"Structure model","details":"Coordinates and associated ncs operations (if present) transformed into standard crystal frame","ordinal":2,"provider":"repository","revision_ordinal":6,"type":"Remediation"}],"pdbx_audit_revision_group":[{"data_content_type":"Structure model","group":"Version format compliance","ordinal":1,"revision_ordinal":2},{"data_content_type":"Structure model","group":"Advisory","ordinal":2,"revision_ordinal":3},{"data_content_type":"Structure model","group":"Version format compliance","ordinal":3,"revision_ordinal":3},{"data_content_type":"Structure model","group":"Advisory","ordinal":4,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Atomic model","ordinal":5,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Data collection","ordinal":6,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Database references","ordinal":7,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Other","ordinal":8,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Source and taxonomy","ordinal":9,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Structure summary","ordinal":10,"revision_ordinal":4},{"data_content_type":"Structure model","group":"Atomic model","ordinal":11,"revision_ordinal":5},{"data_content_type":"Structure model","group":"Data collection","ordinal":12,"revision_ordinal":5},{"data_content_type":"Structure model","group":"Derived calculations","ordinal":13,"revision_ordinal":5},{"data_content_type":"Structure model","group":"Advisory","ordinal":14,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Atomic model","ordinal":15,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Data collection","ordinal":16,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Database references","ordinal":17,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Derived calculations","ordinal":18,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Other","ordinal":19,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Refinement description","ordinal":20,"revision_ordinal":6},{"data_content_type":"Structure model","group":"Advisory","ordinal":21,"revision_ordinal":7}],"pdbx_audit_revision_history":[{"data_content_type":"Structure model","major_revision":1,"minor_revision":0,"ordinal":1,"revision_date":"1984-07-17T00:00:00+0000"},{"data_content_type":"Structure model","major_revision":1,"minor_revision":1,"ordinal":2,"revision_date":"2008-03-03T00:00:00+0000"},{"data_content_type":"Structure model","major_revision":1,"minor_revision":2,"ordinal":3,"revision_date":"2011-07-13T00:00:00+0000"},{"data_content_type":"Structure model","major_revision":2,"minor_revision":0,"ordinal":4,"revision_date":"2020-06-17T00:00:00+0000"},{"data_content_type":"Structure model","major_revision":3,"minor_revision":0,"ordinal":5,"revision_date":"2021-03-31T00:00:00+0000"},{"data_content_type":"Structure model","major_revision":4,"minor_revision":0,"ordinal":6,"revision_date":"2023-02-08T00:00:00+0000"},{"data_content_type":"Structure model","major_revision":4,"minor_revision":1,"ordinal":7,"revision_date":"2023-03-15T00:00:00+0000"}],"pdbx_audit_revision_item":[{"data_content_type":"Structure model","item":"_atom_site.B_iso_or_equiv","ordinal":1,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_atom_site.Cartn_x","ordinal":2,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_atom_site.Cartn_y","ordinal":3,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_atom_site.Cartn_z","ordinal":4,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_entity.pdbx_description","ordinal":5,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_entity_src_gen.gene_src_common_name","ordinal":6,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_entity_src_gen.pdbx_beg_seq_num","ordinal":7,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_entity_src_gen.pdbx_end_seq_num","ordinal":8,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_entity_src_gen.pdbx_gene_src_gene","ordinal":9,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_entity_src_gen.pdbx_seq_type","ordinal":10,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_pdbx_database_status.process_site","ordinal":11,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_pdbx_validate_rmsd_angle.angle_deviation","ordinal":12,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_pdbx_validate_rmsd_angle.angle_value","ordinal":13,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_pdbx_validate_rmsd_bond.bond_deviation","ordinal":14,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_pdbx_validate_rmsd_bond.bond_value","ordinal":15,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_struct_ref.pdbx_align_begin","ordinal":16,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_struct_ref_seq.db_align_beg","ordinal":17,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_struct_ref_seq.db_align_end","ordinal":18,"revision_ordinal":4},{"data_content_type":"Structure model","item":"_atom_site.B_iso_or_equiv","ordinal":19,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_atom_site.Cartn_x","ordinal":20,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_atom_site.Cartn_y","ordinal":21,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_atom_site.Cartn_z","ordinal":22,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_pdbx_validate_rmsd_bond.bond_deviation","ordinal":23,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_pdbx_validate_rmsd_bond.bond_value","ordinal":24,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_struct_site.pdbx_auth_asym_id","ordinal":25,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_struct_site.pdbx_auth_comp_id","ordinal":26,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_struct_site.pdbx_auth_seq_id","ordinal":27,"revision_ordinal":5},{"data_content_type":"Structure model","item":"_atom_site.Cartn_x","ordinal":28,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_site.Cartn_y","ordinal":29,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_site.Cartn_z","ordinal":30,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[1][1]","ordinal":31,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[1][2]","ordinal":32,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[1][3]","ordinal":33,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[2][1]","ordinal":34,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[2][2]","ordinal":35,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[2][3]","ordinal":36,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[3][1]","ordinal":37,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[3][2]","ordinal":38,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_matrix[3][3]","ordinal":39,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_vector[1]","ordinal":40,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_vector[2]","ordinal":41,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_atom_sites.fract_transf_vector[3]","ordinal":42,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_2.pdbx_DOI","ordinal":43,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_2.pdbx_database_accession","ordinal":44,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[1][1]","ordinal":45,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[1][2]","ordinal":46,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[1][3]","ordinal":47,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[2][1]","ordinal":48,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[2][2]","ordinal":49,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[2][3]","ordinal":50,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[3][1]","ordinal":51,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[3][2]","ordinal":52,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx[3][3]","ordinal":53,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx_vector[1]","ordinal":54,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx_vector[2]","ordinal":55,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_database_PDB_matrix.origx_vector[3]","ordinal":56,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_struct_conn_angle.value","ordinal":57,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_validate_close_contact.dist","ordinal":58,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_validate_peptide_omega.omega","ordinal":59,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_validate_planes.rmsd","ordinal":60,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_validate_polymer_linkage.dist","ordinal":61,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_validate_torsion.phi","ordinal":62,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_pdbx_validate_torsion.psi","ordinal":63,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[1][1]","ordinal":64,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[1][2]","ordinal":65,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[1][3]","ordinal":66,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[2][1]","ordinal":67,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[2][2]","ordinal":68,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[2][3]","ordinal":69,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[3][1]","ordinal":70,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[3][2]","ordinal":71,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.matrix[3][3]","ordinal":72,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.vector[1]","ordinal":73,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.vector[2]","ordinal":74,"revision_ordinal":6},{"data_content_type":"Structure model","item":"_struct_ncs_oper.vector[3]","ordinal":75,"revision_ordinal":6}],"pdbx_database_pdbobs_spr":[{"date":"1984-07-17T00:00:00+0000","id":"SPRSDE","pdb_id":"4HHB","replace_pdb_id":"1HHB"}],"pdbx_database_related":[{"content_type":"unspecified","db_id":"2HHB","db_name":"PDB","details":"REFINED BY THE METHOD OF JACK AND LEVITT. THIS\n ENTRY PRESENTS THE BEST ESTIMATE OF THE\n COORDINATES."},{"content_type":"unspecified","db_id":"3HHB","db_name":"PDB","details":"SYMMETRY AVERAGED ABOUT THE (NON-CRYSTALLOGRAPHIC)\n MOLECULAR AXIS AND THEN RE-REGULARIZED BY THE\n ENERGY REFINEMENT METHOD OF LEVITT. THIS ENTRY\n PRESENTS COORDINATES THAT ARE ADEQUATE FOR MOST\n PURPOSES, SUCH AS COMPARISON WITH OTHER STRUCTURES."},{"content_type":"unspecified","db_id":"1GLI","db_name":"PDB"}],"pdbx_database_status":{"pdb_format_compatible":"Y","process_site":"BNL","recvd_initial_deposition_date":"1984-03-07T00:00:00+0000","status_code":"REL"},"rcsb_accession_info":{"deposit_date":"1984-03-07T00:00:00+0000","has_released_experimental_data":"N","initial_release_date":"1984-07-17T00:00:00+0000","major_revision":4,"minor_revision":1,"revision_date":"2023-03-15T00:00:00+0000","status_code":"REL"},"rcsb_entry_container_identifiers":{"assembly_ids":["1"],"entity_ids":["1","2","3","4","5"],"entry_id":"4HHB","model_ids":[1],"non_polymer_entity_ids":["3","4"],"polymer_entity_ids":["1","2"],"rcsb_id":"4HHB","pubmed_id":6726807},"rcsb_entry_info":{"assembly_count":1,"branched_entity_count":0,"cis_peptide_count":0,"deposited_atom_count":4779,"deposited_hydrogen_atom_count":0,"deposited_model_count":1,"deposited_modeled_polymer_monomer_count":574,"deposited_nonpolymer_entity_instance_count":6,"deposited_polymer_entity_instance_count":4,"deposited_polymer_monomer_count":574,"deposited_solvent_atom_count":221,"deposited_unmodeled_polymer_monomer_count":0,"disulfide_bond_count":0,"entity_count":5,"experimental_method":"X-ray","experimental_method_count":1,"inter_mol_covalent_bond_count":0,"inter_mol_metalic_bond_count":4,"molecular_weight":64.74,"na_polymer_entity_types":"Other","nonpolymer_bound_components":["HEM"],"nonpolymer_entity_count":2,"nonpolymer_molecular_weight_maximum":0.62,"nonpolymer_molecular_weight_minimum":0.09,"polymer_composition":"heteromeric protein","polymer_entity_count":2,"polymer_entity_count_dna":0,"polymer_entity_count_rna":0,"polymer_entity_count_nucleic_acid":0,"polymer_entity_count_nucleic_acid_hybrid":0,"polymer_entity_count_protein":2,"polymer_entity_taxonomy_count":2,"polymer_molecular_weight_maximum":15.89,"polymer_molecular_weight_minimum":15.15,"polymer_monomer_count_maximum":146,"polymer_monomer_count_minimum":141,"resolution_combined":[1.74],"selected_polymer_entity_types":"Protein (only)","solvent_entity_count":1,"structure_determination_methodology":"experimental","structure_determination_methodology_priority":10,"diffrn_resolution_high":{"provenance_source":"From refinement resolution cutoff","value":1.74}},"rcsb_primary_citation":{"country":"UK","id":"primary","journal_abbrev":"J.Mol.Biol.","journal_id_astm":"JMOBAK","journal_id_csd":"0070","journal_id_issn":"0022-2836","journal_volume":"175","page_first":"159","page_last":"174","pdbx_database_id_doi":"10.1016/0022-2836(84)90472-8","pdbx_database_id_pub_med":6726807,"rcsb_orcididentifiers":["?","?","?","?"],"rcsb_authors":["Fermi, G.","Perutz, M.F.","Shaanan, B.","Fourme, R."],"rcsb_journal_abbrev":"J Mol Biol","title":"The crystal structure of human deoxyhaemoglobin at 1.74 A resolution","year":1984},"refine":[{"details":"THE COORDINATES GIVEN HERE ARE IN THE ORTHOGONAL ANGSTROM\nSYSTEM STANDARD FOR HEMOGLOBINS. THE Y AXIS IS THE\n(NON CRYSTALLOGRAPHIC) MOLECULAR DIAD AND THE X AXIS IS THE\nPSEUDO DIAD WHICH RELATES THE ALPHA-1 AND BETA-1 CHAINS.\nTHE TRANSFORMATION GIVEN IN THE *MTRIX* RECORDS BELOW\nWILL GENERATE COORDINATES FOR THE *C* AND *D* CHAINS FROM\nTHE *A* AND *B* CHAINS RESPECTIVELY.","ls_rfactor_rwork":0.135,"ls_dres_high":1.74,"pdbx_diffrn_id":["1"],"pdbx_refine_id":"X-RAY DIFFRACTION"}],"refine_hist":[{"cycle_id":"LAST","d_res_high":1.74,"number_atoms_solvent":221,"number_atoms_total":4779,"pdbx_number_atoms_ligand":174,"pdbx_number_atoms_nucleic_acid":0,"pdbx_number_atoms_protein":4384,"pdbx_refine_id":"X-RAY DIFFRACTION"}],"struct":{"title":"THE CRYSTAL STRUCTURE OF HUMAN DEOXYHAEMOGLOBIN AT 1.74 ANGSTROMS RESOLUTION"},"struct_keywords":{"pdbx_keywords":"OXYGEN TRANSPORT","text":"OXYGEN TRANSPORT"},"symmetry":{"int_tables_number":4,"space_group_name_hm":"P 1 21 1"},"rcsb_id":"4HHB"}
pdbinfo = json.loads(response.text)
pdbinfo
{'audit_author': [{'name': 'Fermi, G.', 'pdbx_ordinal': 1},
{'name': 'Perutz, M.F.', 'pdbx_ordinal': 2}],
'cell': {'angle_alpha': 90.0,
'angle_beta': 99.34,
'angle_gamma': 90.0,
'length_a': 63.15,
'length_b': 83.59,
'length_c': 53.8,
'zpdb': 4},
'citation': [{'country': 'UK',
'id': 'primary',
'journal_abbrev': 'J.Mol.Biol.',
'journal_id_astm': 'JMOBAK',
'journal_id_csd': '0070',
'journal_id_issn': '0022-2836',
'journal_volume': '175',
'page_first': '159',
'page_last': '174',
'pdbx_database_id_doi': '10.1016/0022-2836(84)90472-8',
'pdbx_database_id_pub_med': 6726807,
'rcsb_authors': ['Fermi, G.', 'Perutz, M.F.', 'Shaanan, B.', 'Fourme, R.'],
'rcsb_is_primary': 'Y',
'rcsb_journal_abbrev': 'J Mol Biol',
'title': 'The crystal structure of human deoxyhaemoglobin at 1.74 A resolution',
'year': 1984},
{'country': 'UK',
'id': '1',
'journal_abbrev': 'Nature',
'journal_id_astm': 'NATUAS',
'journal_id_csd': '0006',
'journal_id_issn': '0028-0836',
'journal_volume': '295',
'page_first': '535',
'rcsb_authors': ['Perutz, M.F.',
'Hasnain, S.S.',
'Duke, P.J.',
'Sessler, J.L.',
'Hahn, J.E.'],
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'Nature',
'title': 'Stereochemistry of Iron in Deoxyhaemoglobin',
'year': 1982},
{'country': 'US',
'id': '3',
'journal_abbrev': 'Annu.Rev.Biochem.',
'journal_id_astm': 'ARBOAW',
'journal_id_csd': '0413',
'journal_id_issn': '0066-4154',
'journal_volume': '48',
'page_first': '327',
'rcsb_authors': ['Perutz, M.F.'],
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'Annu Rev Biochem',
'title': 'Regulation of Oxygen Affinity of Hemoglobin. Influence of Structure of the Globin on the Heme Iron',
'year': 1979},
{'country': 'UK',
'id': '4',
'journal_abbrev': 'J.Mol.Biol.',
'journal_id_astm': 'JMOBAK',
'journal_id_csd': '0070',
'journal_id_issn': '0022-2836',
'journal_volume': '100',
'page_first': '3',
'rcsb_authors': ['Teneyck, L.F.', 'Arnone, A.'],
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'J Mol Biol',
'title': 'Three-Dimensional Fourier Synthesis of Human Deoxyhemoglobin at 2.5 Angstroms Resolution, I.X-Ray Analysis',
'year': 1976},
{'country': 'UK',
'id': '5',
'journal_abbrev': 'J.Mol.Biol.',
'journal_id_astm': 'JMOBAK',
'journal_id_csd': '0070',
'journal_id_issn': '0022-2836',
'journal_volume': '97',
'page_first': '237',
'rcsb_authors': ['Fermi, G.'],
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'J Mol Biol',
'title': 'Three-Dimensional Fourier Synthesis of Human Deoxyhaemoglobin at 2.5 Angstroms Resolution, Refinement of the Atomic Model',
'year': 1975},
{'country': 'UK',
'id': '6',
'journal_abbrev': 'Nature',
'journal_id_astm': 'NATUAS',
'journal_id_csd': '0006',
'journal_id_issn': '0028-0836',
'journal_volume': '228',
'page_first': '516',
'rcsb_authors': ['Muirhead, H.', 'Greer, J.'],
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'Nature',
'title': 'Three-Dimensional Fourier Synthesis of Human Deoxyhaemoglobin at 3.5 Angstroms Resolution',
'year': 1970},
{'book_publisher': 'Oxford University Press',
'id': '2',
'journal_abbrev': 'Haemoglobin and Myoglobin. Atlas of Molecular Structures in Biology',
'journal_id_csd': '0986',
'journal_id_issn': '0-19-854706-4',
'journal_volume': '2',
'rcsb_authors': ['Fermi, G.', 'Perutz, M.F.'],
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'Haemoglobin And Myoglobin Atlas Of Molecular Structures In Biology',
'year': 1981},
{'book_publisher': 'National Biomedical Research Foundation, Silver Spring,Md.',
'id': '7',
'journal_abbrev': 'Atlas of Protein Sequence and Structure (Data Section)',
'journal_id_csd': '0435',
'journal_id_issn': '0-912466-02-2',
'journal_volume': '5',
'page_first': '56',
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'Atlas Of Protein Sequence And Structure (data Section)',
'year': 1972},
{'book_publisher': 'National Biomedical Research Foundation, Silver Spring,Md.',
'id': '8',
'journal_abbrev': 'Atlas of Protein Sequence and Structure (Data Section)',
'journal_id_csd': '0435',
'journal_id_issn': '0-912466-02-2',
'journal_volume': '5',
'page_first': '64',
'rcsb_is_primary': 'N',
'rcsb_journal_abbrev': 'Atlas Of Protein Sequence And Structure (data Section)',
'year': 1972}],
'diffrn': [{'crystal_id': '1', 'id': '1'}],
'entry': {'id': '4HHB'},
'exptl': [{'method': 'X-RAY DIFFRACTION'}],
'exptl_crystal': [{'density_matthews': 2.26,
'density_percent_sol': 45.48,
'id': '1'}],
'pdbx_audit_revision_category': [{'category': 'atom_site',
'data_content_type': 'Structure model',
'ordinal': 1,
'revision_ordinal': 4},
{'category': 'database_PDB_caveat',
'data_content_type': 'Structure model',
'ordinal': 2,
'revision_ordinal': 4},
{'category': 'entity',
'data_content_type': 'Structure model',
'ordinal': 3,
'revision_ordinal': 4},
{'category': 'entity_name_com',
'data_content_type': 'Structure model',
'ordinal': 4,
'revision_ordinal': 4},
{'category': 'entity_src_gen',
'data_content_type': 'Structure model',
'ordinal': 5,
'revision_ordinal': 4},
{'category': 'pdbx_database_status',
'data_content_type': 'Structure model',
'ordinal': 6,
'revision_ordinal': 4},
{'category': 'pdbx_validate_rmsd_angle',
'data_content_type': 'Structure model',
'ordinal': 7,
'revision_ordinal': 4},
{'category': 'pdbx_validate_rmsd_bond',
'data_content_type': 'Structure model',
'ordinal': 8,
'revision_ordinal': 4},
{'category': 'struct_ref',
'data_content_type': 'Structure model',
'ordinal': 9,
'revision_ordinal': 4},
{'category': 'struct_ref_seq',
'data_content_type': 'Structure model',
'ordinal': 10,
'revision_ordinal': 4},
{'category': 'atom_site',
'data_content_type': 'Structure model',
'ordinal': 11,
'revision_ordinal': 5},
{'category': 'pdbx_validate_rmsd_angle',
'data_content_type': 'Structure model',
'ordinal': 12,
'revision_ordinal': 5},
{'category': 'pdbx_validate_rmsd_bond',
'data_content_type': 'Structure model',
'ordinal': 13,
'revision_ordinal': 5},
{'category': 'struct_site',
'data_content_type': 'Structure model',
'ordinal': 14,
'revision_ordinal': 5},
{'category': 'atom_site',
'data_content_type': 'Structure model',
'ordinal': 15,
'revision_ordinal': 6},
{'category': 'atom_sites',
'data_content_type': 'Structure model',
'ordinal': 16,
'revision_ordinal': 6},
{'category': 'database_2',
'data_content_type': 'Structure model',
'ordinal': 17,
'revision_ordinal': 6},
{'category': 'database_PDB_matrix',
'data_content_type': 'Structure model',
'ordinal': 18,
'revision_ordinal': 6},
{'category': 'pdbx_struct_conn_angle',
'data_content_type': 'Structure model',
'ordinal': 19,
'revision_ordinal': 6},
{'category': 'pdbx_validate_close_contact',
'data_content_type': 'Structure model',
'ordinal': 20,
'revision_ordinal': 6},
{'category': 'pdbx_validate_main_chain_plane',
'data_content_type': 'Structure model',
'ordinal': 21,
'revision_ordinal': 6},
{'category': 'pdbx_validate_peptide_omega',
'data_content_type': 'Structure model',
'ordinal': 22,
'revision_ordinal': 6},
{'category': 'pdbx_validate_planes',
'data_content_type': 'Structure model',
'ordinal': 23,
'revision_ordinal': 6},
{'category': 'pdbx_validate_polymer_linkage',
'data_content_type': 'Structure model',
'ordinal': 24,
'revision_ordinal': 6},
{'category': 'pdbx_validate_rmsd_angle',
'data_content_type': 'Structure model',
'ordinal': 25,
'revision_ordinal': 6},
{'category': 'pdbx_validate_rmsd_bond',
'data_content_type': 'Structure model',
'ordinal': 26,
'revision_ordinal': 6},
{'category': 'pdbx_validate_torsion',
'data_content_type': 'Structure model',
'ordinal': 27,
'revision_ordinal': 6},
{'category': 'struct_ncs_oper',
'data_content_type': 'Structure model',
'ordinal': 28,
'revision_ordinal': 6},
{'category': 'pdbx_database_remark',
'data_content_type': 'Structure model',
'ordinal': 29,
'revision_ordinal': 7}],
'pdbx_audit_revision_details': [{'data_content_type': 'Structure model',
'ordinal': 1,
'provider': 'repository',
'revision_ordinal': 1,
'type': 'Initial release'},
{'data_content_type': 'Structure model',
'details': 'Coordinates and associated ncs operations (if present) transformed into standard crystal frame',
'ordinal': 2,
'provider': 'repository',
'revision_ordinal': 6,
'type': 'Remediation'}],
'pdbx_audit_revision_group': [{'data_content_type': 'Structure model',
'group': 'Version format compliance',
'ordinal': 1,
'revision_ordinal': 2},
{'data_content_type': 'Structure model',
'group': 'Advisory',
'ordinal': 2,
'revision_ordinal': 3},
{'data_content_type': 'Structure model',
'group': 'Version format compliance',
'ordinal': 3,
'revision_ordinal': 3},
{'data_content_type': 'Structure model',
'group': 'Advisory',
'ordinal': 4,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Atomic model',
'ordinal': 5,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Data collection',
'ordinal': 6,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Database references',
'ordinal': 7,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Other',
'ordinal': 8,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Source and taxonomy',
'ordinal': 9,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Structure summary',
'ordinal': 10,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'group': 'Atomic model',
'ordinal': 11,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'group': 'Data collection',
'ordinal': 12,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'group': 'Derived calculations',
'ordinal': 13,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'group': 'Advisory',
'ordinal': 14,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Atomic model',
'ordinal': 15,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Data collection',
'ordinal': 16,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Database references',
'ordinal': 17,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Derived calculations',
'ordinal': 18,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Other',
'ordinal': 19,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Refinement description',
'ordinal': 20,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'group': 'Advisory',
'ordinal': 21,
'revision_ordinal': 7}],
'pdbx_audit_revision_history': [{'data_content_type': 'Structure model',
'major_revision': 1,
'minor_revision': 0,
'ordinal': 1,
'revision_date': '1984-07-17T00:00:00+0000'},
{'data_content_type': 'Structure model',
'major_revision': 1,
'minor_revision': 1,
'ordinal': 2,
'revision_date': '2008-03-03T00:00:00+0000'},
{'data_content_type': 'Structure model',
'major_revision': 1,
'minor_revision': 2,
'ordinal': 3,
'revision_date': '2011-07-13T00:00:00+0000'},
{'data_content_type': 'Structure model',
'major_revision': 2,
'minor_revision': 0,
'ordinal': 4,
'revision_date': '2020-06-17T00:00:00+0000'},
{'data_content_type': 'Structure model',
'major_revision': 3,
'minor_revision': 0,
'ordinal': 5,
'revision_date': '2021-03-31T00:00:00+0000'},
{'data_content_type': 'Structure model',
'major_revision': 4,
'minor_revision': 0,
'ordinal': 6,
'revision_date': '2023-02-08T00:00:00+0000'},
{'data_content_type': 'Structure model',
'major_revision': 4,
'minor_revision': 1,
'ordinal': 7,
'revision_date': '2023-03-15T00:00:00+0000'}],
'pdbx_audit_revision_item': [{'data_content_type': 'Structure model',
'item': '_atom_site.B_iso_or_equiv',
'ordinal': 1,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_x',
'ordinal': 2,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_y',
'ordinal': 3,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_z',
'ordinal': 4,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_entity.pdbx_description',
'ordinal': 5,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_entity_src_gen.gene_src_common_name',
'ordinal': 6,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_entity_src_gen.pdbx_beg_seq_num',
'ordinal': 7,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_entity_src_gen.pdbx_end_seq_num',
'ordinal': 8,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_entity_src_gen.pdbx_gene_src_gene',
'ordinal': 9,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_entity_src_gen.pdbx_seq_type',
'ordinal': 10,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_pdbx_database_status.process_site',
'ordinal': 11,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_rmsd_angle.angle_deviation',
'ordinal': 12,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_rmsd_angle.angle_value',
'ordinal': 13,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_rmsd_bond.bond_deviation',
'ordinal': 14,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_rmsd_bond.bond_value',
'ordinal': 15,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_struct_ref.pdbx_align_begin',
'ordinal': 16,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_struct_ref_seq.db_align_beg',
'ordinal': 17,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_struct_ref_seq.db_align_end',
'ordinal': 18,
'revision_ordinal': 4},
{'data_content_type': 'Structure model',
'item': '_atom_site.B_iso_or_equiv',
'ordinal': 19,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_x',
'ordinal': 20,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_y',
'ordinal': 21,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_z',
'ordinal': 22,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_rmsd_bond.bond_deviation',
'ordinal': 23,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_rmsd_bond.bond_value',
'ordinal': 24,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_struct_site.pdbx_auth_asym_id',
'ordinal': 25,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_struct_site.pdbx_auth_comp_id',
'ordinal': 26,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_struct_site.pdbx_auth_seq_id',
'ordinal': 27,
'revision_ordinal': 5},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_x',
'ordinal': 28,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_y',
'ordinal': 29,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_site.Cartn_z',
'ordinal': 30,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[1][1]',
'ordinal': 31,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[1][2]',
'ordinal': 32,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[1][3]',
'ordinal': 33,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[2][1]',
'ordinal': 34,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[2][2]',
'ordinal': 35,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[2][3]',
'ordinal': 36,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[3][1]',
'ordinal': 37,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[3][2]',
'ordinal': 38,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_matrix[3][3]',
'ordinal': 39,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_vector[1]',
'ordinal': 40,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_vector[2]',
'ordinal': 41,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_atom_sites.fract_transf_vector[3]',
'ordinal': 42,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_2.pdbx_DOI',
'ordinal': 43,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_2.pdbx_database_accession',
'ordinal': 44,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[1][1]',
'ordinal': 45,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[1][2]',
'ordinal': 46,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[1][3]',
'ordinal': 47,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[2][1]',
'ordinal': 48,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[2][2]',
'ordinal': 49,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[2][3]',
'ordinal': 50,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[3][1]',
'ordinal': 51,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[3][2]',
'ordinal': 52,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx[3][3]',
'ordinal': 53,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx_vector[1]',
'ordinal': 54,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx_vector[2]',
'ordinal': 55,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_database_PDB_matrix.origx_vector[3]',
'ordinal': 56,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_struct_conn_angle.value',
'ordinal': 57,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_close_contact.dist',
'ordinal': 58,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_peptide_omega.omega',
'ordinal': 59,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_planes.rmsd',
'ordinal': 60,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_polymer_linkage.dist',
'ordinal': 61,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_torsion.phi',
'ordinal': 62,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_pdbx_validate_torsion.psi',
'ordinal': 63,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[1][1]',
'ordinal': 64,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[1][2]',
'ordinal': 65,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[1][3]',
'ordinal': 66,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[2][1]',
'ordinal': 67,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[2][2]',
'ordinal': 68,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[2][3]',
'ordinal': 69,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[3][1]',
'ordinal': 70,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[3][2]',
'ordinal': 71,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.matrix[3][3]',
'ordinal': 72,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.vector[1]',
'ordinal': 73,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.vector[2]',
'ordinal': 74,
'revision_ordinal': 6},
{'data_content_type': 'Structure model',
'item': '_struct_ncs_oper.vector[3]',
'ordinal': 75,
'revision_ordinal': 6}],
'pdbx_database_pdbobs_spr': [{'date': '1984-07-17T00:00:00+0000',
'id': 'SPRSDE',
'pdb_id': '4HHB',
'replace_pdb_id': '1HHB'}],
'pdbx_database_related': [{'content_type': 'unspecified',
'db_id': '2HHB',
'db_name': 'PDB',
'details': 'REFINED BY THE METHOD OF JACK AND LEVITT. THIS\n ENTRY PRESENTS THE BEST ESTIMATE OF THE\n COORDINATES.'},
{'content_type': 'unspecified',
'db_id': '3HHB',
'db_name': 'PDB',
'details': 'SYMMETRY AVERAGED ABOUT THE (NON-CRYSTALLOGRAPHIC)\n MOLECULAR AXIS AND THEN RE-REGULARIZED BY THE\n ENERGY REFINEMENT METHOD OF LEVITT. THIS ENTRY\n PRESENTS COORDINATES THAT ARE ADEQUATE FOR MOST\n PURPOSES, SUCH AS COMPARISON WITH OTHER STRUCTURES.'},
{'content_type': 'unspecified', 'db_id': '1GLI', 'db_name': 'PDB'}],
'pdbx_database_status': {'pdb_format_compatible': 'Y',
'process_site': 'BNL',
'recvd_initial_deposition_date': '1984-03-07T00:00:00+0000',
'status_code': 'REL'},
'rcsb_accession_info': {'deposit_date': '1984-03-07T00:00:00+0000',
'has_released_experimental_data': 'N',
'initial_release_date': '1984-07-17T00:00:00+0000',
'major_revision': 4,
'minor_revision': 1,
'revision_date': '2023-03-15T00:00:00+0000',
'status_code': 'REL'},
'rcsb_entry_container_identifiers': {'assembly_ids': ['1'],
'entity_ids': ['1', '2', '3', '4', '5'],
'entry_id': '4HHB',
'model_ids': [1],
'non_polymer_entity_ids': ['3', '4'],
'polymer_entity_ids': ['1', '2'],
'rcsb_id': '4HHB',
'pubmed_id': 6726807},
'rcsb_entry_info': {'assembly_count': 1,
'branched_entity_count': 0,
'cis_peptide_count': 0,
'deposited_atom_count': 4779,
'deposited_hydrogen_atom_count': 0,
'deposited_model_count': 1,
'deposited_modeled_polymer_monomer_count': 574,
'deposited_nonpolymer_entity_instance_count': 6,
'deposited_polymer_entity_instance_count': 4,
'deposited_polymer_monomer_count': 574,
'deposited_solvent_atom_count': 221,
'deposited_unmodeled_polymer_monomer_count': 0,
'disulfide_bond_count': 0,
'entity_count': 5,
'experimental_method': 'X-ray',
'experimental_method_count': 1,
'inter_mol_covalent_bond_count': 0,
'inter_mol_metalic_bond_count': 4,
'molecular_weight': 64.74,
'na_polymer_entity_types': 'Other',
'nonpolymer_bound_components': ['HEM'],
'nonpolymer_entity_count': 2,
'nonpolymer_molecular_weight_maximum': 0.62,
'nonpolymer_molecular_weight_minimum': 0.09,
'polymer_composition': 'heteromeric protein',
'polymer_entity_count': 2,
'polymer_entity_count_dna': 0,
'polymer_entity_count_rna': 0,
'polymer_entity_count_nucleic_acid': 0,
'polymer_entity_count_nucleic_acid_hybrid': 0,
'polymer_entity_count_protein': 2,
'polymer_entity_taxonomy_count': 2,
'polymer_molecular_weight_maximum': 15.89,
'polymer_molecular_weight_minimum': 15.15,
'polymer_monomer_count_maximum': 146,
'polymer_monomer_count_minimum': 141,
'resolution_combined': [1.74],
'selected_polymer_entity_types': 'Protein (only)',
'solvent_entity_count': 1,
'structure_determination_methodology': 'experimental',
'structure_determination_methodology_priority': 10,
'diffrn_resolution_high': {'provenance_source': 'From refinement resolution cutoff',
'value': 1.74}},
'rcsb_primary_citation': {'country': 'UK',
'id': 'primary',
'journal_abbrev': 'J.Mol.Biol.',
'journal_id_astm': 'JMOBAK',
'journal_id_csd': '0070',
'journal_id_issn': '0022-2836',
'journal_volume': '175',
'page_first': '159',
'page_last': '174',
'pdbx_database_id_doi': '10.1016/0022-2836(84)90472-8',
'pdbx_database_id_pub_med': 6726807,
'rcsb_orcididentifiers': ['?', '?', '?', '?'],
'rcsb_authors': ['Fermi, G.', 'Perutz, M.F.', 'Shaanan, B.', 'Fourme, R.'],
'rcsb_journal_abbrev': 'J Mol Biol',
'title': 'The crystal structure of human deoxyhaemoglobin at 1.74 A resolution',
'year': 1984},
'refine': [{'details': 'THE COORDINATES GIVEN HERE ARE IN THE ORTHOGONAL ANGSTROM\nSYSTEM STANDARD FOR HEMOGLOBINS. THE Y AXIS IS THE\n(NON CRYSTALLOGRAPHIC) MOLECULAR DIAD AND THE X AXIS IS THE\nPSEUDO DIAD WHICH RELATES THE ALPHA-1 AND BETA-1 CHAINS.\nTHE TRANSFORMATION GIVEN IN THE *MTRIX* RECORDS BELOW\nWILL GENERATE COORDINATES FOR THE *C* AND *D* CHAINS FROM\nTHE *A* AND *B* CHAINS RESPECTIVELY.',
'ls_rfactor_rwork': 0.135,
'ls_dres_high': 1.74,
'pdbx_diffrn_id': ['1'],
'pdbx_refine_id': 'X-RAY DIFFRACTION'}],
'refine_hist': [{'cycle_id': 'LAST',
'd_res_high': 1.74,
'number_atoms_solvent': 221,
'number_atoms_total': 4779,
'pdbx_number_atoms_ligand': 174,
'pdbx_number_atoms_nucleic_acid': 0,
'pdbx_number_atoms_protein': 4384,
'pdbx_refine_id': 'X-RAY DIFFRACTION'}],
'struct': {'title': 'THE CRYSTAL STRUCTURE OF HUMAN DEOXYHAEMOGLOBIN AT 1.74 ANGSTROMS RESOLUTION'},
'struct_keywords': {'pdbx_keywords': 'OXYGEN TRANSPORT',
'text': 'OXYGEN TRANSPORT'},
'symmetry': {'int_tables_number': 4, 'space_group_name_hm': 'P 1 21 1'},
'rcsb_id': '4HHB'}
pdbinfo['rcsb_entry_info']
{'assembly_count': 1,
'branched_entity_count': 0,
'cis_peptide_count': 0,
'deposited_atom_count': 4779,
'deposited_hydrogen_atom_count': 0,
'deposited_model_count': 1,
'deposited_modeled_polymer_monomer_count': 574,
'deposited_nonpolymer_entity_instance_count': 6,
'deposited_polymer_entity_instance_count': 4,
'deposited_polymer_monomer_count': 574,
'deposited_solvent_atom_count': 221,
'deposited_unmodeled_polymer_monomer_count': 0,
'disulfide_bond_count': 0,
'entity_count': 5,
'experimental_method': 'X-ray',
'experimental_method_count': 1,
'inter_mol_covalent_bond_count': 0,
'inter_mol_metalic_bond_count': 4,
'molecular_weight': 64.74,
'na_polymer_entity_types': 'Other',
'nonpolymer_bound_components': ['HEM'],
'nonpolymer_entity_count': 2,
'nonpolymer_molecular_weight_maximum': 0.62,
'nonpolymer_molecular_weight_minimum': 0.09,
'polymer_composition': 'heteromeric protein',
'polymer_entity_count': 2,
'polymer_entity_count_dna': 0,
'polymer_entity_count_rna': 0,
'polymer_entity_count_nucleic_acid': 0,
'polymer_entity_count_nucleic_acid_hybrid': 0,
'polymer_entity_count_protein': 2,
'polymer_entity_taxonomy_count': 2,
'polymer_molecular_weight_maximum': 15.89,
'polymer_molecular_weight_minimum': 15.15,
'polymer_monomer_count_maximum': 146,
'polymer_monomer_count_minimum': 141,
'resolution_combined': [1.74],
'selected_polymer_entity_types': 'Protein (only)',
'solvent_entity_count': 1,
'structure_determination_methodology': 'experimental',
'structure_determination_methodology_priority': 10,
'diffrn_resolution_high': {'provenance_source': 'From refinement resolution cutoff',
'value': 1.74}}
response = requests.get('https://data.rcsb.org/rest/v1/core/polymer_entity/4hhb/1')
json.loads(response.text)
{'rcsb_cluster_membership': [{'cluster_id': 105, 'identity': 100},
{'cluster_id': 112, 'identity': 95},
{'cluster_id': 96, 'identity': 90},
{'cluster_id': 47, 'identity': 70},
{'cluster_id': 20, 'identity': 50},
{'cluster_id': 31, 'identity': 30}],
'entity_poly': {'nstd_linkage': 'no',
'nstd_monomer': 'no',
'pdbx_seq_one_letter_code': 'VLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPNALSALSDLHAHKLRVDPVNFKLLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTVLTSKYR',
'pdbx_seq_one_letter_code_can': 'VLSPADKTNVKAAWGKVGAHAGEYGAEALERMFLSFPTTKTYFPHFDLSHGSAQVKGHGKKVADALTNAVAHVDDMPNALSALSDLHAHKLRVDPVNFKLLSHCLLVTLAAHLPAEFTPAVHASLDKFLASVSTVLTSKYR',
'pdbx_strand_id': 'A,C',
'rcsb_artifact_monomer_count': 0,
'rcsb_conflict_count': 0,
'rcsb_deletion_count': 0,
'rcsb_entity_polymer_type': 'Protein',
'rcsb_insertion_count': 0,
'rcsb_mutation_count': 0,
'rcsb_non_std_monomer_count': 0,
'rcsb_sample_sequence_length': 141,
'type': 'polypeptide(L)'},
'entity_src_gen': [{'gene_src_common_name': 'Human',
'gene_src_genus': 'Homo',
'pdbx_alt_source_flag': 'sample',
'pdbx_beg_seq_num': 1,
'pdbx_end_seq_num': 141,
'pdbx_gene_src_gene': 'HBA1, HBA2',
'pdbx_gene_src_ncbi_taxonomy_id': '9606',
'pdbx_gene_src_scientific_name': 'Homo sapiens',
'pdbx_seq_type': 'Biological sequence',
'pdbx_src_id': 1}],
'rcsb_entity_host_organism': [{'beg_seq_num': 1,
'end_seq_num': 141,
'pdbx_src_id': '1',
'provenance_source': 'Primary Data'}],
'rcsb_entity_source_organism': [{'beg_seq_num': 1,
'common_name': 'Human',
'end_seq_num': 141,
'ncbi_common_names': ['human'],
'ncbi_parent_scientific_name': 'Eukaryota',
'ncbi_scientific_name': 'Homo sapiens',
'ncbi_taxonomy_id': 9606,
'pdbx_src_id': '1',
'provenance_source': 'Primary Data',
'scientific_name': 'Homo sapiens',
'source_type': 'genetically engineered',
'taxonomy_lineage': [{'depth': 1,
'id': '131567',
'name': 'cellular organisms'},
{'depth': 1, 'id': '131567', 'name': 'biota'},
{'depth': 2, 'id': '2759', 'name': 'Eukaryota'},
{'depth': 2, 'id': '2759', 'name': 'Eucarya'},
{'depth': 2, 'id': '2759', 'name': 'Eucaryotae'},
{'depth': 2, 'id': '2759', 'name': 'Eukarya'},
{'depth': 2, 'id': '2759', 'name': 'Eukaryotae'},
{'depth': 2, 'id': '2759', 'name': 'eucaryotes'},
{'depth': 2, 'id': '2759', 'name': 'eukaryotes'},
{'depth': 3, 'id': '33154', 'name': 'Opisthokonta'},
{'depth': 3, 'id': '33154', 'name': 'Fungi/Metazoa group'},
{'depth': 3, 'id': '33154', 'name': 'opisthokonts'},
{'depth': 4, 'id': '33208', 'name': 'Metazoa'},
{'depth': 4, 'id': '33208', 'name': 'Animalia'},
{'depth': 4, 'id': '33208', 'name': 'metazoans'},
{'depth': 4, 'id': '33208', 'name': 'multicellular animals'},
{'depth': 5, 'id': '6072', 'name': 'Eumetazoa'},
{'depth': 6, 'id': '33213', 'name': 'Bilateria'},
{'depth': 7, 'id': '33511', 'name': 'Deuterostomia'},
{'depth': 7, 'id': '33511', 'name': 'deuterostomes'},
{'depth': 8, 'id': '7711', 'name': 'Chordata'},
{'depth': 8, 'id': '7711', 'name': 'chordates'},
{'depth': 9, 'id': '89593', 'name': 'Craniata'},
{'depth': 10, 'id': '7742', 'name': 'Vertebrata'},
{'depth': 10, 'id': '7742', 'name': 'vertebrates'},
{'depth': 11, 'id': '7776', 'name': 'Gnathostomata'},
{'depth': 11, 'id': '7776', 'name': 'jawed vertebrates'},
{'depth': 12, 'id': '117570', 'name': 'Teleostomi'},
{'depth': 13, 'id': '117571', 'name': 'Euteleostomi'},
{'depth': 13, 'id': '117571', 'name': 'bony vertebrates'},
{'depth': 14, 'id': '8287', 'name': 'Sarcopterygii'},
{'depth': 15, 'id': '1338369', 'name': 'Dipnotetrapodomorpha'},
{'depth': 16, 'id': '32523', 'name': 'Tetrapoda'},
{'depth': 16, 'id': '32523', 'name': 'tetrapods'},
{'depth': 17, 'id': '32524', 'name': 'Amniota'},
{'depth': 17, 'id': '32524', 'name': 'amniotes'},
{'depth': 18, 'id': '40674', 'name': 'Mammalia'},
{'depth': 18, 'id': '40674', 'name': 'mammals'},
{'depth': 19, 'id': '32525', 'name': 'Theria'},
{'depth': 20, 'id': '9347', 'name': 'Eutheria'},
{'depth': 20, 'id': '9347', 'name': 'Placentalia'},
{'depth': 20, 'id': '9347', 'name': 'eutherian mammals'},
{'depth': 20, 'id': '9347', 'name': 'placental mammals'},
{'depth': 20, 'id': '9347', 'name': 'placentals'},
{'depth': 21, 'id': '1437010', 'name': 'Boreoeutheria'},
{'depth': 21, 'id': '1437010', 'name': 'Boreotheria'},
{'depth': 22, 'id': '314146', 'name': 'Euarchontoglires'},
{'depth': 23, 'id': '9443', 'name': 'Primates'},
{'depth': 23, 'id': '9443', 'name': 'Primata'},
{'depth': 23, 'id': '9443', 'name': 'primate'},
{'depth': 24, 'id': '376913', 'name': 'Haplorrhini'},
{'depth': 25, 'id': '314293', 'name': 'Simiiformes'},
{'depth': 25, 'id': '314293', 'name': 'Anthropoidea'},
{'depth': 26, 'id': '9526', 'name': 'Catarrhini'},
{'depth': 27, 'id': '314295', 'name': 'Hominoidea'},
{'depth': 27, 'id': '314295', 'name': 'ape'},
{'depth': 27, 'id': '314295', 'name': 'apes'},
{'depth': 28, 'id': '9604', 'name': 'Hominidae'},
{'depth': 28, 'id': '9604', 'name': 'Pongidae'},
{'depth': 28, 'id': '9604', 'name': 'great apes'},
{'depth': 29, 'id': '207598', 'name': 'Homininae'},
{'depth': 29, 'id': '207598', 'name': 'Homo/Pan/Gorilla group'},
{'depth': 30, 'id': '9605', 'name': 'Homo'},
{'depth': 30, 'id': '9605', 'name': 'humans'},
{'depth': 31, 'id': '9606', 'name': 'Homo sapiens'},
{'depth': 31, 'id': '9606', 'name': 'human'}],
'rcsb_gene_name': [{'provenance_source': 'Primary Data', 'value': 'HBA1'},
{'provenance_source': 'Primary Data', 'value': 'HBA2'},
{'provenance_source': 'UniProt', 'value': 'HBA1'},
{'provenance_source': 'UniProt', 'value': 'HBA2'}]}],
'rcsb_polymer_entity': {'formula_weight': 15.15,
'pdbx_description': 'Hemoglobin subunit alpha',
'pdbx_number_of_molecules': 2,
'rcsb_multiple_source_flag': 'N',
'rcsb_source_part_count': 1,
'rcsb_source_taxonomy_count': 1,
'src_method': 'man',
'rcsb_macromolecular_names_combined': [{'name': 'Hemoglobin subunit alpha',
'provenance_code': 'ECO:0000304',
'provenance_source': 'PDB Preferred Name'},
{'name': 'Alpha-globin',
'provenance_code': 'ECO:0000303',
'provenance_source': 'PDB Synonym'},
{'name': 'Hemoglobin alpha chain',
'provenance_code': 'ECO:0000303',
'provenance_source': 'PDB Synonym'}],
'rcsb_polymer_name_combined': {'names': ['Hemoglobin subunit alpha'],
'provenance_source': 'UniProt Name'}},
'rcsb_polymer_entity_align': [{'provenance_source': 'SIFTS',
'reference_database_accession': 'P69905',
'reference_database_name': 'UniProt',
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 141,
'ref_beg_seq_id': 2}]}],
'rcsb_polymer_entity_annotation': [{'annotation_id': 'PF00042',
'assignment_version': '34.0',
'name': 'Globin (Globin)',
'provenance_source': 'Pfam',
'type': 'Pfam'},
{'annotation_id': 'P69905',
'assignment_version': '1.0',
'name': 'Glycoprotein',
'provenance_source': 'PDB',
'type': 'GlyGen'}],
'rcsb_polymer_entity_container_identifiers': {'asym_ids': ['A', 'C'],
'auth_asym_ids': ['A', 'C'],
'chem_comp_monomers': ['ALA',
'ARG',
'ASN',
'ASP',
'CYS',
'GLN',
'GLU',
'GLY',
'HIS',
'LEU',
'LYS',
'MET',
'PHE',
'PRO',
'SER',
'THR',
'TRP',
'TYR',
'VAL'],
'entity_id': '1',
'entry_id': '4HHB',
'rcsb_id': '4HHB_1',
'reference_sequence_identifiers': [{'database_accession': 'P69905',
'database_name': 'UniProt',
'entity_sequence_coverage': 1.0,
'provenance_source': 'SIFTS',
'reference_sequence_coverage': 0.9929577464788732}],
'uniprot_ids': ['P69905']},
'rcsb_polymer_entity_feature': [{'assignment_version': '34.0',
'feature_id': 'PF00042',
'name': 'Globin (Globin)',
'provenance_source': 'Pfam',
'type': 'Pfam',
'feature_positions': [{'beg_seq_id': 26, 'end_seq_id': 136}]},
{'name': 'Hydropathy values',
'provenance_source': 'biojava-7.0.1',
'type': 'hydropathy',
'feature_positions': [{'beg_seq_id': 5,
'values': [-0.47,
-0.47,
-1.32,
-1.03,
-0.66,
-0.96,
-0.61,
-0.61,
-0.07,
0.28,
0.01,
0.09,
0.09,
-0.16,
-0.44,
-0.54,
-0.16,
-0.42,
-0.77,
-0.77,
0.01,
-0.58,
-1.03,
-0.43,
0.02,
0.49,
0.2,
0.9,
0.52,
0.02,
0.33,
0.4,
0.11,
-0.34,
-0.46,
-0.54,
-1.21,
-0.72,
-1.03,
-0.53,
-0.19,
-0.47,
-0.37,
-0.77,
-0.39,
-0.42,
-0.27,
-0.31,
-0.78,
-1.04,
-0.73,
-1.12,
-1.47,
-1.2,
-0.61,
-1.47,
-0.83,
-0.37,
-0.09,
-0.43,
0.2,
1.1,
0.83,
0.28,
1.13,
0.54,
-0.27,
0.02,
0.23,
-0.36,
-0.62,
-0.4,
-0.13,
-0.4,
0.41,
0.71,
0.11,
0.71,
0.74,
0.74,
-0.03,
-0.38,
-0.16,
-1.08,
-0.52,
-0.52,
-1.12,
-0.3,
-0.89,
-0.22,
-0.22,
-0.22,
0.7,
0.14,
0.18,
0.63,
0.59,
1.4,
1.56,
1.91,
1.91,
1.69,
1.98,
1.98,
2.12,
1.52,
1.3,
0.44,
0.83,
0.33,
-0.04,
-0.04,
0.78,
0.0,
0.38,
0.09,
0.9,
0.2,
-0.16,
0.33,
0.56,
0.29,
0.56,
0.82,
0.82,
0.32,
1.18,
2.03,
1.64,
1.13,
0.5,
0.44,
-0.52]}]},
{'name': 'Disordered binding sites',
'provenance_source': 'Anchor2',
'type': 'disorder_binding',
'feature_positions': [{'beg_seq_id': 1,
'values': [0.39,
0.38,
0.37,
0.36,
0.36,
0.36,
0.35,
0.35,
0.35,
0.35,
0.36,
0.37,
0.38,
0.39,
0.39,
0.39,
0.39,
0.39,
0.4,
0.4,
0.39,
0.39,
0.4,
0.41,
0.42,
0.42,
0.4,
0.39,
0.37,
0.36,
0.36,
0.35,
0.35,
0.35,
0.36,
0.37,
0.38,
0.39,
0.4,
0.41,
0.41,
0.41,
0.4,
0.39,
0.39,
0.38,
0.38,
0.38,
0.38,
0.38,
0.39,
0.4,
0.4,
0.41,
0.42,
0.43,
0.42,
0.42,
0.4,
0.4,
0.39,
0.38,
0.38,
0.37,
0.36,
0.36,
0.36,
0.36,
0.36,
0.36,
0.36,
0.37,
0.37,
0.37,
0.38,
0.39,
0.39,
0.4,
0.41,
0.42,
0.42,
0.43,
0.43,
0.42,
0.41,
0.4,
0.4,
0.41,
0.41,
0.42,
0.43,
0.42,
0.4,
0.38,
0.37,
0.35,
0.34,
0.32,
0.32,
0.31,
0.31,
0.31,
0.31,
0.3,
0.3,
0.29,
0.28,
0.26,
0.25,
0.23,
0.21,
0.2,
0.19,
0.17,
0.15,
0.14,
0.13,
0.12,
0.12,
0.11,
0.11,
0.11,
0.1,
0.09,
0.09,
0.09,
0.09,
0.08,
0.08,
0.08,
0.07,
0.07,
0.07,
0.07,
0.07,
0.08,
0.08,
0.08,
0.08,
0.08,
0.09]}]},
{'name': 'Disordered regions',
'provenance_source': 'IUPred2(short)',
'type': 'disorder',
'feature_positions': [{'beg_seq_id': 1,
'values': [0.9,
0.85,
0.79,
0.68,
0.64,
0.6,
0.5,
0.46,
0.38,
0.34,
0.27,
0.31,
0.37,
0.28,
0.26,
0.26,
0.27,
0.24,
0.18,
0.19,
0.26,
0.23,
0.18,
0.16,
0.21,
0.15,
0.17,
0.22,
0.22,
0.26,
0.25,
0.21,
0.14,
0.15,
0.24,
0.16,
0.19,
0.12,
0.13,
0.19,
0.17,
0.17,
0.18,
0.27,
0.28,
0.29,
0.37,
0.35,
0.35,
0.35,
0.35,
0.31,
0.36,
0.44,
0.38,
0.31,
0.38,
0.39,
0.44,
0.39,
0.37,
0.38,
0.32,
0.35,
0.33,
0.35,
0.38,
0.39,
0.36,
0.28,
0.29,
0.26,
0.22,
0.24,
0.24,
0.18,
0.25,
0.21,
0.2,
0.23,
0.18,
0.21,
0.14,
0.18,
0.21,
0.15,
0.18,
0.08,
0.08,
0.05,
0.05,
0.05,
0.06,
0.06,
0.03,
0.02,
0.02,
0.02,
0.01,
0.01,
0.01,
0.01,
0.01,
0.01,
0.01,
0.01,
0.01,
0.01,
0.02,
0.01,
0.01,
0.02,
0.02,
0.02,
0.01,
0.03,
0.05,
0.04,
0.03,
0.04,
0.04,
0.03,
0.03,
0.06,
0.03,
0.03,
0.02,
0.05,
0.05,
0.03,
0.03,
0.07,
0.1,
0.16,
0.2,
0.34,
0.38,
0.41,
0.57,
0.69,
0.76]}]}],
'rcsb_polymer_entity_feature_summary': [{'count': 0,
'coverage': 0.0,
'type': 'CARD_MODEL'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_DESCRIPTION'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_DOMAIN_NAME'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_GENE_ALLELE_NAME'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_ORGANISM_NAME'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_PROTEIN_NAME'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_RECEPTOR_DESCRIPTION'},
{'count': 0, 'coverage': 0.0, 'type': 'IMGT_ANTIBODY_RECEPTOR_TYPE'},
{'count': 1, 'coverage': 0.78723, 'type': 'Pfam'},
{'count': 0, 'coverage': 0.0, 'type': 'SABDAB_ANTIBODY_ANTIGEN_NAME'},
{'count': 0, 'coverage': 0.0, 'type': 'SABDAB_ANTIBODY_NAME'},
{'count': 0, 'coverage': 0.0, 'type': 'SABDAB_ANTIBODY_TARGET'},
{'count': 0, 'coverage': 0.0, 'type': 'artifact'},
{'count': 0, 'coverage': 0.0, 'type': 'modified_monomer'},
{'count': 0, 'coverage': 0.0, 'type': 'mutation'}],
'rcsb_polymer_entity_name_com': [{'name': 'Alpha-globin,Hemoglobin alpha chain'}],
'rcsb_related_target_references': [{'related_resource_name': 'ChEMBL',
'related_resource_version': '33',
'related_target_id': 'CHEMBL2887',
'target_taxonomy_id': 9606,
'aligned_target': [{'entity_beg_seq_id': 1,
'length': 140,
'target_beg_seq_id': 2}]},
{'related_resource_name': 'ChEMBL',
'related_resource_version': '33',
'related_target_id': 'CHEMBL2095168',
'target_taxonomy_id': 9606,
'aligned_target': [{'entity_beg_seq_id': 1,
'length': 140,
'target_beg_seq_id': 2}]},
{'related_resource_name': 'DrugBank',
'related_resource_version': '5.1',
'related_target_id': 'P69905',
'target_taxonomy_id': 9606,
'aligned_target': [{'entity_beg_seq_id': 1,
'length': 140,
'target_beg_seq_id': 2}]},
{'related_resource_name': 'Pharos',
'related_resource_version': '6.13.4',
'related_target_id': '6231',
'target_taxonomy_id': 9606,
'aligned_target': [{'entity_beg_seq_id': 1,
'length': 140,
'target_beg_seq_id': 2}]}],
'rcsb_target_cofactors': [{'cofactor_name': 'Iron Dextran',
'cofactor_resource_id': 'DB00893',
'mechanism_of_action': 'After iron dextran is injected, the circulating iron dextran is removed from the plasma by cells of the reticuloendothelial system, which split the complex into its components of iron and dextran. The iron is immediately bound to the available protein moieties to form hemosiderin or ferritin, the physiological forms of iron, or to a lesser extent to transferrin. This iron which is subject to physiological control replenishes hemoglobin and depleted iron stores.',
'neighbor_flag': 'N',
'pubmed_ids': [17139284, 17016423, 11752352],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'XEEYBQQBJWHFJM-UHFFFAOYSA-N',
'cofactor_smiles': '[Fe]',
'cofactor_name': 'Iron',
'cofactor_resource_id': 'DB01592',
'mechanism_of_action': 'Iron is necessary for the production of hemoglobin. Iron-deficiency can lead to decreased production of hemoglobin and a microcytic, hypochromic anemia.',
'neighbor_flag': 'N',
'pubmed_ids': [16901899],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'HCHKCACWOHOZIP-UHFFFAOYSA-N',
'cofactor_smiles': '[Zn]',
'cofactor_name': 'Zinc',
'cofactor_resource_id': 'DB01593',
'mechanism_of_action': '**Zinc has three primary biological roles**: _catalytic_, _structural_, and _regulatory_. The catalytic and structural role of zinc is well established, and there are various noteworthy reviews on these functions. For example, zinc is a structural constituent in numerous proteins, inclusive of growth factors, cytokines, receptors, enzymes, and transcription factors for different cellular signaling pathways. It is implicated in numerous cellular processes as a cofactor for approximately 3000 human proteins including enzymes, nuclear factors, and hormones [L2096].\r\n\r\nZinc promotes resistance to epithelial apoptosis through cell protection (cytoprotection) against reactive oxygen species and bacterial toxins, likely through the antioxidant activity of the cysteine-rich metallothioneins [A32419].\r\n\r\nIn HL-60 cells (promyelocytic leukemia cell line), zinc enhances the up-regulation of A20 mRNA, which, via TRAF pathway, decreases NF-kappaB activation, leading to decreased gene expression and generation of tumor necrosis factor-alpha (TNF-alpha), IL-1beta, and IL-8 [A32418].\r\n\r\nThere are several mechanisms of action of zinc on acute diarrhea. Various mechanisms are specific to the gastrointestinal system: zinc restores mucosal barrier integrity and enterocyte brush-border enzyme activity, it promotes the production of antibodies and circulating lymphocytes against intestinal pathogens, and has a direct effect on ion channels, acting as a potassium channel blocker of adenosine 3-5-cyclic monophosphate-mediated chlorine secretion. Cochrane researchers examined the evidence available up to 30 September 2016 [L2106].\r\n\r\nZinc deficiency in humans decreases the activity of serum _thymulin_ (a hormone of the thymus), which is necessary for the maturation of T-helper cells. T-helper 1 (Th(1)) cytokines are decreased but T-helper 2 (Th(2)) cytokines are not affected by zinc deficiency in humans [A32417].\r\n\r\nThe change of _Th(1)_ to _Th(2)_ function leads to cell-mediated immune dysfunction. Because IL-2 production (Th(1) cytokine) is decreased, this causes decreased activity of natural-killer-cell (NK cell) and T cytolytic cells, normally involved in killing viruses, bacteria, and malignant cells [A32424]. \r\n\r\nIn humans, zinc deficiency may lead to the generation of new CD4+ T cells, produced in the thymus. In cell culture studies (HUT-78, a Th(0) human malignant lymphoblastoid cell line), as a result of zinc deficiency, nuclear factor-kappaB (NF-kappaB) activation, phosphorylation of IkappaB, and binding of NF-kappaB to DNA are decreased and this results in decreased Th(1) cytokine production [A32417].\r\n\r\nIn another study, zinc supplementation in human subjects suppressed the gene expression and production of pro-inflammatory cytokines and decreased oxidative stress markers [A32424]. In HL-60 cells (a human pro-myelocytic leukemia cell line), zinc deficiency increased the levels of TNF-alpha, IL-1beta, and IL-8 cytokines and mRNA. In such cells, zinc was found to induce A20, a zinc finger protein that inhibited NF-kappaB activation by the tumor necrosis factor receptor-associated factor pathway. This process decreased gene expression of pro-inflammatory cytokines and oxidative stress markers [A32417].\r\n\r\nThe exact mechanism of zinc in acne treatment is poorly understood. However, zinc is considered to act directly on microbial inflammatory equilibrium and facilitate antibiotic absorption when used in combination with other agents. Topical zinc alone as well as in combination with other agents may be efficacious because of its anti-inflammatory activity and ability to reduce P. acnes bacteria by the inhibition of P. acnes lipases and free fatty acid levels [L2102].',
'neighbor_flag': 'N',
'pubmed_ids': [23896426],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'HAEJSGLKJYIYTB-ZZXKWVIFSA-N',
'cofactor_smiles': 'OC(=O)\\C=C\\C1=CC=C(C=C1)C(O)=O',
'cofactor_chem_comp_id': 'CIN',
'cofactor_name': '4-Carboxycinnamic Acid',
'cofactor_resource_id': 'DB02126',
'neighbor_flag': 'N',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'TZRXHJWUDPFEEY-UHFFFAOYSA-N',
'cofactor_smiles': '[O-][N+](=O)OCC(CO[N+]([O-])=O)(CO[N+]([O-])=O)CO[N+]([O-])=O',
'cofactor_name': 'Pentaerythritol tetranitrate',
'cofactor_resource_id': 'DB06154',
'mechanism_of_action': 'Pentaerythritol tetranitrate is the lipid soluble polyol ester of nitric acid belonging to the family of _nitro-vasodilators_. Pentaerythritol tetranitrate releases free nitric oxide (NO) after the denitration reaction, which triggers NO-dependent signaling transduction involving soluble _guanylate cyclase (sGC_). Nitric oxide binds reversibly to the ferrous-heme center of sGC, causing conformational change and activating the enzyme. This enzyme activation results in increased cellular concentrations of _cyclic guanosine monophosphate _(cGMP) within the vascular smooth muscle, resulting in vasodilation mediated by cGMP-dependent protein kinases. Additionally, this agent causes dose-dependent arterial and venous bed [L2393].',
'neighbor_flag': 'N',
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'AVXQTLSOXWQOHO-UHFFFAOYSA-N',
'cofactor_smiles': 'COC1=C(OCC2=NC=CC=C2)C=C(C)C=C1',
'cofactor_chem_comp_id': 'B77',
'cofactor_name': '2-[(2-methoxy-5-methylphenoxy)methyl]pyridine',
'cofactor_resource_id': 'DB07427',
'neighbor_flag': 'Y',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'MBHBRRBLXCXQKV-UHFFFAOYSA-N',
'cofactor_smiles': 'COC1=CC(OCC2=CC=NC=C2)=C(C)C=C1',
'cofactor_chem_comp_id': 'B78',
'cofactor_name': '4-[(5-methoxy-2-methylphenoxy)methyl]pyridine',
'cofactor_resource_id': 'DB07428',
'neighbor_flag': 'Y',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'CXMXRPHRNRROMY-UHFFFAOYSA-N',
'cofactor_smiles': 'OC(=O)CCCCCCCCC(O)=O',
'cofactor_chem_comp_id': 'DEC',
'cofactor_name': 'Sebacic acid',
'cofactor_resource_id': 'DB07645',
'neighbor_flag': 'N',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'OYJPTSMWFKGZJM-UHFFFAOYSA-N',
'cofactor_smiles': 'CC(C)(OC1=CC=C(NC(=O)NC2=CC(Cl)=CC(Cl)=C2)C=C1)C(O)=O',
'cofactor_chem_comp_id': 'L35',
'cofactor_name': '2-[4-({[(3,5-DICHLOROPHENYL)AMINO]CARBONYL}AMINO)PHENOXY]-2-METHYLPROPANOIC ACID',
'cofactor_resource_id': 'DB08077',
'neighbor_flag': 'Y',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'RXOHFPCZGPKIRD-UHFFFAOYSA-N',
'cofactor_smiles': 'OC(=O)C1=CC2=CC=C(C=C2C=C1)C(O)=O',
'cofactor_chem_comp_id': 'NDD',
'cofactor_name': '2,6-dicarboxynaphthalene',
'cofactor_resource_id': 'DB08262',
'neighbor_flag': 'N',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'BNFRJXLZYUTIII-UHFFFAOYSA-N',
'cofactor_smiles': 'CC1=CC(NC(=O)CC2=CC=C(OC(C)(C)C(O)=O)C=C2)=CC(C)=C1',
'cofactor_chem_comp_id': 'RQ3',
'cofactor_name': 'Efaproxiral',
'cofactor_resource_id': 'DB08486',
'neighbor_flag': 'Y',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'QMKYBPDZANOJGF-UHFFFAOYSA-N',
'cofactor_smiles': 'OC(=O)C1=CC(=CC(=C1)C(O)=O)C(O)=O',
'cofactor_chem_comp_id': 'TMM',
'cofactor_name': 'Trimesic acid',
'cofactor_resource_id': 'DB08632',
'neighbor_flag': 'N',
'pubmed_ids': [10592235],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'IOVCWXUNBOPUCH-UHFFFAOYSA-N',
'cofactor_smiles': 'ON=O',
'cofactor_chem_comp_id': '2NO',
'cofactor_name': 'Nitrous acid',
'cofactor_resource_id': 'DB09112',
'mechanism_of_action': 'Cyanide has a high affinity for the oxidized form of iron (Fe3+) such as that found in cytochrome oxidase a3 [A19441]. Cyanide binds to and inhibits cytochrome oxidase a3, preventing oxidative phophorylation from occuring. The resultant lack of ATP cannot support normal cellular processes, particularly in the brain. Compensatory increases in anaerobic respiration result in rising levels of lactic acid and subsequent acidosis. \r\n\r\nNitrite primarily acts by oxidizing hemoglobin to methemoglobin [A19440]. The now oxidized Fe3+ in methemoglobin also binds cyanide with high affinity and accepts cyanide from cytochrome a3. This leaves cytochrome a3 free to resume its function in oxidative phosphorylation. The slow dissociation of cyanide from methemoglobin allows hepatic enzymes such as rhodanese to detoxify the compound without further systemic toxicity occuring. Methemoglobin is reduced back to hemoglobin by methemoglobin reductase allowing the affected blood cells to resume normal functioning.\r\n\r\nThe reduction of nitrite by hemoglobin results in the formation of nitric oxide [A19442]. Nitric oxide acts as a powerful vasodilator, producing vascular smooth muscle relaxation through activation of soluble guanylate cyclase and the subsequent cyclic guanylyl triphosphate mediated signalling cascade [A19443].',
'neighbor_flag': 'N',
'pubmed_ids': [1569239],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'RYGMFSIKBFXOCR-UHFFFAOYSA-N',
'cofactor_smiles': '[Cu]',
'cofactor_name': 'Copper',
'cofactor_resource_id': 'DB09130',
'mechanism_of_action': "Copper is absorbed from the gut via high affinity copper uptake protein and likely through low affinity copper uptake protein and natural resistance-associated macrophage protein-2 [A19528]. It is believed that copper is reduced to the Cu1+ form prior to transport. Once inside the enterocyte, it is bound to copper transport protein ATOX1 which shuttles the ion to copper transporting ATPase-1 on the golgi membrane which take up copper into the golgi apparatus. Once copper has been secreted by enterocytes into the systemic circulation it remain largely bound by ceruloplasmin (65-90%), albumin (18%), and alpha 2-macroglobulin (12%). \r\n\r\nCopper is an essential element in the body and is incorporated into many oxidase enzymes as a cofactor [A19518]. It is also a component of zinc/copper super oxide dismutase, giving it an anti-oxidant role. Copper defiency occurs in Occipital Horn Syndrome and Menke's disease both of which are associated with impaired development of connective tissue due to the lack of copper to act as a cofactor in protein-lysine-6-oxidase. Menke's disease is also associated with progressive neurological impairment leading to death in infancy. The precise mechanisms of the effects of copper deficiency are vague due to the wide range of enzymes which use the ion as a cofactor.\r\n\r\nCopper appears to reduce the viabilty and motility of spermatozoa [A19526]. This reduces the likelihood of fertilization with a copper IUD, producing copper's contraceptive effect [A19526]. The exact mechanism of copper's effect on sperm are unknown.",
'neighbor_flag': 'N',
'pubmed_ids': [23896426],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'MYMOFIZGZYHOMD-UHFFFAOYSA-N',
'cofactor_smiles': 'O=O',
'cofactor_chem_comp_id': 'OXY',
'cofactor_name': 'Oxygen',
'cofactor_resource_id': 'DB09140',
'mechanism_of_action': 'Oxygen therapy increases the arterial pressure of oxygen and is effective in improving gas exchange and oxygen delivery to tissues, provided that there are functional alveolar units. Oxygen plays a critical role as an electron acceptor during oxidative phosphorylation in the electron transport chain through activation of cytochrome c oxidase (terminal enzyme of the electron transport chain). This process achieves successful aerobic respiration in organisms to generate ATP molecules as an energy source in many tissues. Oxygen supplementation acts to restore normal cellular activity at the mitochondrial level and reduce metabolic acidosis. There is also evidence that oxygen may interact with O2-sensitive voltage-gated potassium channels in glomus cells and cause hyperpolarization of mitochondrial membrane [A19120]. ',
'neighbor_flag': 'Y',
'pubmed_ids': [1634355],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'CADNYOZXMIKYPR-UHFFFAOYSA-B',
'cofactor_smiles': '[Fe+3].[Fe+3].[Fe+3].[Fe+3].[O-]P([O-])(=O)OP([O-])([O-])=O.[O-]P([O-])(=O)OP([O-])([O-])=O.[O-]P([O-])(=O)OP([O-])([O-])=O',
'cofactor_name': 'Ferric pyrophosphate',
'cofactor_resource_id': 'DB09147',
'mechanism_of_action': 'The usage of ferric pyrophosphate is based on the strong complex formation between these two species. Besides, the capacity of pyrophosphate to trigger iron removal from transferrin, enhance iron transfer from transferrin to ferritin and promote iron exchange between transferrin molecules. These properties make it a very suitable compound for parenteral administration, iron delivery into circulation and incorporation into hemoglobin.[A31979]',
'neighbor_flag': 'N',
'pubmed_ids': [10231452],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'MQBDAEHWGRMADS-XNHLMZCASA-M',
'cofactor_smiles': '[O--].[O--].[O--].[Na+].[Fe+3].[Fe+3].OC[C@@H](O)[C@@H](O)[C@H](O)[C@@H](O)C([O-])=O.OC[C@H]1O[C@@](CO)(O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@@H](O)[C@@H]1O.OC[C@H]1O[C@@](CO)(O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@@H](O)[C@@H]1O.OC[C@H]1O[C@@](CO)(O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@@H](O)[C@@H]1O.OC[C@H]1O[C@@](CO)(O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@@H](O)[C@@H]1O.OC[C@H]1O[C@@](CO)(O[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@@H](O)[C@@H]1O',
'cofactor_name': 'Sodium ferric gluconate complex',
'cofactor_resource_id': 'DB09517',
'mechanism_of_action': 'The complex is endocytosed by macrophages of the reticuloendothelial system. Within an endosome of the macrophage , lysosome fuses with the endosome creating an acidic environment leading to the cleavage of the complex from iron. Iron is then incorporated in ferritin, transferrin or hemoglobin. Sodium ferric gluconate also normalizes RBC production by binding with hemoglobin ',
'neighbor_flag': 'N',
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'BAUYGSIQEAFULO-UHFFFAOYSA-L',
'cofactor_smiles': '[Fe++].[O-]S([O-])(=O)=O',
'cofactor_name': 'Ferrous sulfate anhydrous',
'cofactor_resource_id': 'DB13257',
'mechanism_of_action': 'Iron is required to maintain optimal health, particularly for helping to form red blood cells (RBC) that carry oxygen around the body. A deficiency in iron indicates that the body cannot produce enough normal red blood cells.[A32514,L11800] Iron deficiency anemia occurs when body stores of iron decrease to very low levels, and the stored iron is insufficient to support normal red blood cell (RBC) production. Insufficient dietary iron, impaired iron absorption, bleeding, pregnancy, or loss of iron through the urine can lead to iron deficiency.[A32514,L11794] Symptoms of iron deficiency anemia include fatigue, breathlessness, palpitations, dizziness, and headache.\r\n\r\nTaking iron in supplement form, such as ferrous sulfate, allows for more rapid increases in iron levels when dietary supply and stores are not sufficient.[L2175] Iron is transported by the divalent metal transporter 1 (DMT1) across the endolysosomal membrane to enter the macrophage. It can then can be incorporated into ferritin and be stored in the macrophage or carried of the macrophage by ferroportin. This exported iron is oxidized by the enzyme to ceruloplasmin to Fe3+, followed by sequestration by transferrin for transport in the serum to various sites, including the bone marrow for hemoglobin synthesis or into the liver.[A32524] Iron combines with porphyrin and globin chains to form hemoglobin, which is critical for oxygen delivery from the lungs to other tissues.[L2263]',
'neighbor_flag': 'N',
'pubmed_ids': [24310424, 21694802, 18954837],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'SXAWSYZURCZSDX-UHFFFAOYSA-B',
'cofactor_smiles': '[Fe+3].[Fe+3].[Fe+3].[Fe+3].OP(O)(=O)OP(O)(O)=O.OP(O)(=O)OP(O)(O)=O.OP([O-])(=O)OP([O-])([O-])=O.OC(CC([O-])=O)(CC([O-])=O)C([O-])=O.OC(CC([O-])=O)(CC([O-])=O)C([O-])=O.OC(CC([O-])=O)(CC([O-])=O)C([O-])=O',
'cofactor_name': 'Ferric pyrophosphate citrate',
'cofactor_resource_id': 'DB13995',
'mechanism_of_action': 'The usage of ferric pyrophosphate is based on the strong complex formation between these two species. Besides, the capacity of pyrophosphate to trigger iron removal from transferrin, enhance iron transfer from transferrin to ferritin and promote iron exchange between transferrin molecules. These properties make it a very suitable compound for parenteral administration, iron delivery into circulation and incorporation into hemoglobin.[A31979]',
'neighbor_flag': 'N',
'pubmed_ids': [10231452],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'DJWUNCQRNNEAKC-UHFFFAOYSA-L',
'cofactor_smiles': '[Zn++].CC([O-])=O.CC([O-])=O',
'cofactor_name': 'Zinc acetate',
'cofactor_resource_id': 'DB14487',
'mechanism_of_action': '**Zinc has three primary biological roles**: _catalytic_, _structural_, and _regulatory_. The catalytic and structural role of zinc is well established, and there are various noteworthy reviews on these functions. For example, zinc is a structural constituent in numerous proteins, inclusive of growth factors, cytokines, receptors, enzymes, and transcription factors for different cellular signaling pathways. It is implicated in numerous cellular processes as a cofactor for approximately 3000 human proteins including enzymes, nuclear factors, and hormones [L2096].\r\n\r\nZinc promotes resistance to epithelial apoptosis through cell protection (cytoprotection) against reactive oxygen species and bacterial toxins, likely through the antioxidant activity of the cysteine-rich metallothioneins [A32419].\r\n\r\nIn HL-60 cells (promyelocytic leukemia cell line), zinc enhances the up-regulation of A20 mRNA, which, via TRAF pathway, decreases NF-kappaB activation, leading to decreased gene expression and generation of tumor necrosis factor-alpha (TNF-alpha), IL-1beta, and IL-8 [A32418].\r\n\r\nThere are several mechanisms of action of zinc on acute diarrhea. Various mechanisms are specific to the gastrointestinal system: zinc restores mucosal barrier integrity and enterocyte brush-border enzyme activity, it promotes the production of antibodies and circulating lymphocytes against intestinal pathogens, and has a direct effect on ion channels, acting as a potassium channel blocker of adenosine 3-5-cyclic monophosphate-mediated chlorine secretion. Cochrane researchers examined the evidence available up to 30 September 2016 [L2106].\r\n\r\nZinc deficiency in humans decreases the activity of serum _thymulin_ (a hormone of the thymus), which is necessary for the maturation of T-helper cells. T-helper 1 (Th(1)) cytokines are decreased but T-helper 2 (Th(2)) cytokines are not affected by zinc deficiency in humans [A32417].\r\n\r\nThe change of _Th(1)_ to _Th(2)_ function leads to cell-mediated immune dysfunction. Because IL-2 production (Th(1) cytokine) is decreased, this causes decreased activity of natural-killer-cell (NK cell) and T cytolytic cells, normally involved in killing viruses, bacteria, and malignant cells [A32417]. \r\n\r\nIn humans, zinc deficiency may lead to the generation of new CD4+ T cells, produced in the thymus. In cell culture studies (HUT-78, a Th(0) human malignant lymphoblastoid cell line), as a result of zinc deficiency, nuclear factor-kappaB (NF-kappaB) activation, phosphorylation of IkappaB, and binding of NF-kappaB to DNA are decreased and this results in decreased Th(1) cytokine production [A32417].\r\n\r\nIn another study, zinc supplementation in human subjects suppressed the gene expression and production of pro-inflammatory cytokines and decreased oxidative stress markers [A32417]. In HL-60 cells (a human pro-myelocytic leukemia cell line), zinc deficiency increased the levels of TNF-alpha, IL-1beta, and IL-8 cytokines and mRNA. In such cells, zinc was found to induce A20, a zinc finger protein that inhibited NF-kappaB activation by the tumor necrosis factor receptor-associated factor pathway. This process decreased gene expression of pro-inflammatory cytokines and oxidative stress markers [A32417].\r\n\r\nThe exact mechanism of zinc in acne treatment is poorly understood. However, zinc is considered to act directly on microbial inflammatory equilibrium and facilitate antibiotic absorption when used in combination with other agents. Topical zinc alone as well as in combination with other agents may be efficacious because of its anti-inflammatory activity and ability to reduce P. acnes bacteria by the inhibition of P. acnes lipases and free fatty acid levels [L2102].',
'neighbor_flag': 'N',
'pubmed_ids': [23896426],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'VRIVJOXICYMTAG-IYEMJOQQSA-L',
'cofactor_smiles': '[Fe++].[H][C@@](O)(CO)[C@@]([H])(O)[C@]([H])(O)[C@@]([H])(O)C([O-])=O.[H][C@@](O)(CO)[C@@]([H])(O)[C@]([H])(O)[C@@]([H])(O)C([O-])=O',
'cofactor_name': 'Ferrous gluconate',
'cofactor_resource_id': 'DB14488',
'mechanism_of_action': 'Iron is necessary for the production of hemoglobin. Iron-deficiency can lead to decreased production of hemoglobin and a microcytic, hypochromic anemia.',
'neighbor_flag': 'N',
'pubmed_ids': [16901899],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'MDXRFOWKIZPNTA-UHFFFAOYSA-L',
'cofactor_smiles': '[Fe++].[O-]C(=O)CCC([O-])=O',
'cofactor_name': 'Ferrous succinate',
'cofactor_resource_id': 'DB14489',
'mechanism_of_action': 'Iron is necessary for the production of hemoglobin. Iron-deficiency can lead to decreased production of hemoglobin and a microcytic, hypochromic anemia.',
'neighbor_flag': 'N',
'pubmed_ids': [16901899],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'RFBYLSCVRUTUSB-ZZMNMWMASA-L',
'cofactor_smiles': '[Fe++].[H][C@](O)(CO)[C@@]1([H])OC(=O)C(O)=C1O.[H][C@](O)(CO)[C@@]1([H])OC(=O)C([O-])=C1[O-]',
'cofactor_name': 'Ferrous ascorbate',
'cofactor_resource_id': 'DB14490',
'mechanism_of_action': 'Iron is necessary for the production of hemoglobin. Iron-deficiency can lead to decreased production of hemoglobin and a microcytic, hypochromic anemia.',
'neighbor_flag': 'N',
'pubmed_ids': [16901899],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'PMVSDNDAUGGCCE-TYYBGVCCSA-L',
'cofactor_smiles': '[Fe++].[H]\\C(=C(\\[H])C([O-])=O)C([O-])=O',
'cofactor_name': 'Ferrous fumarate',
'cofactor_resource_id': 'DB14491',
'mechanism_of_action': 'Iron is necessary for the production of hemoglobin. Iron-deficiency can lead to decreased production of hemoglobin and a microcytic, hypochromic anemia.',
'neighbor_flag': 'N',
'pubmed_ids': [16901899],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'YJYOLOWXCPIBSY-UHFFFAOYSA-L',
'cofactor_smiles': '[Fe++].NCC([O-])=O.NCC([O-])=O.OS(O)(=O)=O',
'cofactor_name': 'Ferrous glycine sulfate',
'cofactor_resource_id': 'DB14501',
'mechanism_of_action': 'Iron is necessary for the production of hemoglobin. Iron-deficiency can lead to decreased production of hemoglobin and a microcytic, hypochromic anemia.',
'neighbor_flag': 'N',
'pubmed_ids': [16901899],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'JIAARYAFYJHUJI-UHFFFAOYSA-L',
'cofactor_smiles': '[Cl-].[Cl-].[Zn++]',
'cofactor_name': 'Zinc chloride',
'cofactor_resource_id': 'DB14533',
'mechanism_of_action': 'Zinc performs catalytic, structural, and regulatory roles in the body. Zinc is a component of approximately 3000 human proteins.[A204104]\r\n\r\nZinc is cytoprotective against reactive oxygen species mediated apoptosis through the action of metallothioneins.[A32419]\r\n\r\nIn a promyelocytic leukemia cell line, zinc enhances the up-regulation of A20 mRNA, which, via the TRAF pathway, decreases NF-kappaB activation, leading to decreased gene expression and generation of TNF-α, IL-1β, and IL-8 [A32418].\r\n\r\nIn patients with diarrhea, zinc restores mucosal barrier integrity, restores enterocyte brush-border enzyme activity, promotes the production of antibodies, and promotes the production of circulating lymphocytes against intestinal pathogens.[A204101] Zinc also directly affects ion channels as a potassium channel blocker of cAMP-mediated chlorine secretion.[A204101]\r\n\r\nZinc deficiency decreases thymulin, inhibiting T-helper cell maturation and decreased Th-1 cytokines like IL-2.[A32417] Decreased IL-2 decreases the activity of NK cells and CD8+ T cells.[A32417] Zinc deficiency also leads to the generation of CD4+ T cells, decreased NF-κB activation, decreased phosphorylation of IκB, and decreased binding of NF-κB to DNA.[A32417]',
'neighbor_flag': 'N',
'pubmed_ids': [646791],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_name': 'Zinc sulfate, unspecified form',
'cofactor_resource_id': 'DB14548',
'neighbor_flag': 'N',
'pubmed_ids': [646791],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'FWCVZAQENIZVMY-UHFFFAOYSA-N',
'cofactor_smiles': 'CC(C)N1N=CC=C1C1=C(COC2=CC=CC(O)=C2C=O)C=CC=N1',
'cofactor_name': 'Voxelotor',
'cofactor_resource_id': 'DB14975',
'mechanism_of_action': 'Deoxygenated sickle hemoglobin (HbS) polymerization is the causal factor for sickle cell disease. The genetic mutation associated with this disease leads to the formation of abnormal, sickle shaped red blood cells that aggregate and block blood vessels throughout the body, causing vaso-occlusive crises.[T734] Voxelotor binds irreversibly with the N‐terminal valine of the α‐chain of hemoglobin, leading to an allosteric modification of Hb20, which increases the affinity for oxygen. Oxygenated HbS does not polymerize.[A188126,A188129] By directly blocking HbS polymerization, voxelotor can successfully treat sickle cell disease by preventing the formation of abnormally shaped cells, which eventually cause lack of oxygenation and blood flow to organs.[A188123,A188138,T734]',
'neighbor_flag': 'N',
'pubmed_ids': [30655275],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'cofactor_in_ch_ikey': 'JTQTXQSGPZRXJF-DOJSGGEQSA-N',
'cofactor_smiles': '[Fe+3].OC[C@H](O)[C@@H](O)[C@H](O)[C@H](O)CO[C@H]1O[C@H](CO[C@H]2O[C@H](CO)[C@@H](O)[C@H](O)[C@H]2O)[C@@H](O)[C@H](O)[C@H]1O',
'cofactor_name': 'Ferric derisomaltose',
'cofactor_resource_id': 'DB15617',
'mechanism_of_action': 'This drug is a complex made of iron (III) hydroxide and derisomaltose, which is an iron carbohydrate oligosaccharide that works to releases iron. The released iron then binds to the transport protein, transferrin, and is taken to erythroid precursor cells[A190528] for incorporation into the hemoglobin molecule.[L11581,L11617]',
'neighbor_flag': 'N',
'pubmed_ids': [29261547, 24310424],
'resource_name': 'DrugBank',
'resource_version': '5.1',
'target_resource_id': 'P69905'},
{'binding_assay_value': 5.09,
'binding_assay_value_type': 'pEC50',
'cofactor_smiles': 'CC(C)N1N=CC=C1C1=C(COC2=C(C=O)C(O)=CC=C2)C=CC=N1',
'cofactor_resource_id': 'CHEMBL4101807',
'mechanism_of_action': 'Voxelotor is a hemoglobin S (HbS) polymerization inhibitor that binds to HbS with a 1:1 stoichiometry and exhibits preferential partitioning to red blood cells (RBCs). By increasing the affinity of Hb for oxygen, voxelotor demonstrates dose-dependent inhibition of HbS polymerization. Nonclinical studies suggest that voxelotor may inhibit RBC sickling, improve RBC deformability, and reduce whole blood viscosity.',
'neighbor_flag': 'N',
'resource_name': 'Pharos',
'resource_version': '6.13.4',
'target_resource_id': '6231'}],
'rcsb_id': '4HHB_1',
'rcsb_polymer_entity_group_membership': [{'group_id': '47_70',
'aggregation_method': 'sequence_identity',
'similarity_cutoff': 70.0,
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 46,
'ref_beg_seq_id': 5},
{'entity_beg_seq_id': 47, 'length': 69, 'ref_beg_seq_id': 52},
{'entity_beg_seq_id': 116, 'length': 26, 'ref_beg_seq_id': 122}]},
{'group_id': '31_30',
'aggregation_method': 'sequence_identity',
'similarity_cutoff': 30.0,
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 31,
'ref_beg_seq_id': 31},
{'entity_beg_seq_id': 32, 'length': 15, 'ref_beg_seq_id': 64},
{'entity_beg_seq_id': 47, 'length': 4, 'ref_beg_seq_id': 81},
{'entity_beg_seq_id': 51, 'length': 10, 'ref_beg_seq_id': 90},
{'entity_beg_seq_id': 61, 'length': 41, 'ref_beg_seq_id': 101},
{'entity_beg_seq_id': 102, 'length': 14, 'ref_beg_seq_id': 143},
{'entity_beg_seq_id': 116, 'length': 26, 'ref_beg_seq_id': 158}]},
{'group_id': 'P69905',
'aggregation_method': 'matching_uniprot_accession',
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 141,
'ref_beg_seq_id': 2}]},
{'group_id': '20_50',
'aggregation_method': 'sequence_identity',
'similarity_cutoff': 50.0,
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 46,
'ref_beg_seq_id': 5},
{'entity_beg_seq_id': 47, 'length': 4, 'ref_beg_seq_id': 52},
{'entity_beg_seq_id': 51, 'length': 51, 'ref_beg_seq_id': 61},
{'entity_beg_seq_id': 102, 'length': 14, 'ref_beg_seq_id': 113},
{'entity_beg_seq_id': 116, 'length': 26, 'ref_beg_seq_id': 128}]},
{'group_id': '96_90',
'aggregation_method': 'sequence_identity',
'similarity_cutoff': 90.0,
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 141,
'ref_beg_seq_id': 5}]},
{'group_id': '105_100',
'aggregation_method': 'sequence_identity',
'similarity_cutoff': 100.0,
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 141,
'ref_beg_seq_id': 5}]},
{'group_id': '112_95',
'aggregation_method': 'sequence_identity',
'similarity_cutoff': 95.0,
'aligned_regions': [{'entity_beg_seq_id': 1,
'length': 141,
'ref_beg_seq_id': 5}]}],
'rcsb_genomic_lineage': [{'id': '9606', 'name': 'Homo sapiens', 'depth': 0},
{'id': '9606:16', 'name': 'Chromosome 16', 'depth': 1},
{'id': '9606:16:hemoglobin_subunit_alpha_2',
'name': 'hemoglobin subunit alpha 2',
'depth': 2},
{'id': '9606:16:hemoglobin_subunit_alpha_1',
'name': 'hemoglobin subunit alpha 1',
'depth': 2}],
'rcsb_cluster_flexibility': {'link': 'http://pdbflex.org/cluster.html#!/1babA/252/4hhbA',
'label': 'Low',
'avg_rmsd': 0.738,
'max_rmsd': 4.392,
'provenance_code': 'PDBFlex'},
'rcsb_latest_revision': {'major_revision': 4, 'minor_revision': 1}}
response = requests.get('https://data.rcsb.org/rest/v1/core/polymer_entity_instance/4HHB/A')
json.loads(response.text)
{'rcsb_ligand_neighbors': [{'atom_id': 'NE2',
'auth_seq_id': 87,
'comp_id': 'HIS',
'distance': 2.143,
'ligand_asym_id': 'E',
'ligand_atom_id': 'FE',
'ligand_comp_id': 'HEM',
'ligand_entity_id': '3',
'ligand_is_bound': 'Y',
'ligand_model_id': 1,
'seq_id': 87}],
'rcsb_polymer_entity_instance_container_identifiers': {'asym_id': 'A',
'auth_asym_id': 'A',
'auth_to_entity_poly_seq_mapping': ['1',
'2',
'3',
'4',
'5',
'6',
'7',
'8',
'9',
'10',
'11',
'12',
'13',
'14',
'15',
'16',
'17',
'18',
'19',
'20',
'21',
'22',
'23',
'24',
'25',
'26',
'27',
'28',
'29',
'30',
'31',
'32',
'33',
'34',
'35',
'36',
'37',
'38',
'39',
'40',
'41',
'42',
'43',
'44',
'45',
'46',
'47',
'48',
'49',
'50',
'51',
'52',
'53',
'54',
'55',
'56',
'57',
'58',
'59',
'60',
'61',
'62',
'63',
'64',
'65',
'66',
'67',
'68',
'69',
'70',
'71',
'72',
'73',
'74',
'75',
'76',
'77',
'78',
'79',
'80',
'81',
'82',
'83',
'84',
'85',
'86',
'87',
'88',
'89',
'90',
'91',
'92',
'93',
'94',
'95',
'96',
'97',
'98',
'99',
'100',
'101',
'102',
'103',
'104',
'105',
'106',
'107',
'108',
'109',
'110',
'111',
'112',
'113',
'114',
'115',
'116',
'117',
'118',
'119',
'120',
'121',
'122',
'123',
'124',
'125',
'126',
'127',
'128',
'129',
'130',
'131',
'132',
'133',
'134',
'135',
'136',
'137',
'138',
'139',
'140',
'141'],
'entity_id': '1',
'entry_id': '4HHB',
'rcsb_id': '4HHB.A'},
'rcsb_polymer_instance_annotation': [{'annotation_id': '1.10.490.10',
'assignment_version': 'v4_2_0',
'name': 'Globins',
'ordinal': 1,
'provenance_source': 'CATH',
'type': 'CATH',
'annotation_lineage': [{'depth': 1, 'id': '1', 'name': 'Mainly Alpha'},
{'depth': 2, 'id': '1.10', 'name': 'Orthogonal Bundle'},
{'depth': 3, 'id': '1.10.490', 'name': 'Globin-like'},
{'depth': 4, 'id': '1.10.490.10', 'name': 'Globins'}]},
{'annotation_id': 'd4hhba_',
'assignment_version': '2.08-stable',
'name': 'Hemoglobin, alpha-chain',
'ordinal': 5,
'provenance_source': 'SCOPe',
'type': 'SCOP',
'annotation_lineage': [{'depth': 1,
'id': '46456',
'name': 'All alpha proteins'},
{'depth': 2, 'id': '46457', 'name': 'Globin-like'},
{'depth': 3, 'id': '46458', 'name': 'Globin-like'},
{'depth': 4, 'id': '46463', 'name': 'Globins'},
{'depth': 5, 'id': '46486', 'name': 'Hemoglobin, alpha-chain'}]},
{'annotation_id': 'e4hhbA1',
'assignment_version': '1.6',
'name': 'Globin',
'ordinal': 9,
'provenance_source': 'ECOD',
'type': 'ECOD',
'annotation_lineage': [{'depth': 1,
'id': '100006',
'name': 'A: alpha arrays'},
{'depth': 2, 'id': '200282', 'name': 'X: Globin-like (From Topology)'},
{'depth': 3, 'id': '300615', 'name': 'H: Globin-like (From Topology)'},
{'depth': 4, 'id': '400688', 'name': 'T: Globin-like'},
{'depth': 5, 'id': '504248', 'name': 'F: Globin'}]}],
'rcsb_polymer_instance_feature': [{'assignment_version': 'v4_2_0',
'feature_id': '1.10.490.10',
'name': 'Globins',
'ordinal': 1,
'provenance_source': 'CATH',
'type': 'CATH',
'feature_positions': [{'beg_seq_id': 1, 'end_seq_id': 141}],
'additional_properties': [{'name': 'CATH_NAME', 'values': ['Globins']},
{'name': 'CATH_DOMAIN_ID', 'values': ['4hhbA00']}]},
{'assignment_version': '2.08-stable',
'feature_id': 'd4hhba_',
'name': 'Hemoglobin, alpha-chain',
'ordinal': 5,
'provenance_source': 'SCOPe',
'type': 'SCOP',
'feature_positions': [{'beg_seq_id': 1, 'end_seq_id': 141}],
'additional_properties': [{'name': 'SCOP_NAME',
'values': ['Hemoglobin', ' alpha-chain']},
{'name': 'SCOP_DOMAIN_ID', 'values': ['d4hhba_']},
{'name': 'SCOP_SUN_ID', 'values': ['46486']}]},
{'assignment_version': '1.6',
'feature_id': 'e4hhbA1',
'name': 'Globin',
'ordinal': 9,
'provenance_source': 'ECOD',
'type': 'ECOD',
'feature_positions': [{'beg_seq_id': 1, 'end_seq_id': 141}],
'additional_properties': [{'name': 'ECOD_FAMILY_NAME',
'values': ['Globin']},
{'name': 'ECOD_DOMAIN_ID', 'values': ['e4hhbA1']}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P1',
'name': 'helix',
'ordinal': 13,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 3, 'end_seq_id': 18}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P2',
'name': 'helix',
'ordinal': 14,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 20, 'end_seq_id': 35}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P3',
'name': 'helix',
'ordinal': 15,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 36, 'end_seq_id': 42}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P4',
'name': 'helix',
'ordinal': 16,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 50, 'end_seq_id': 51}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P5',
'name': 'helix',
'ordinal': 17,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 52, 'end_seq_id': 71}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P6',
'name': 'helix',
'ordinal': 18,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 80, 'end_seq_id': 88}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P7',
'name': 'helix',
'ordinal': 19,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 94, 'end_seq_id': 112}]},
{'assignment_version': 'V1.0',
'feature_id': 'HELX_P8',
'name': 'helix',
'ordinal': 20,
'provenance_source': 'PROMOTIF',
'type': 'HELIX_P',
'feature_positions': [{'beg_seq_id': 118, 'end_seq_id': 138}]},
{'assignment_version': 'V1.0',
'feature_id': '1',
'name': 'unassigned secondary structure',
'ordinal': 45,
'provenance_source': 'PROMOTIF',
'type': 'UNASSIGNED_SEC_STRUCT',
'feature_positions': [{'beg_seq_id': 1, 'end_seq_id': 2},
{'beg_seq_id': 19, 'end_seq_id': 19},
{'beg_seq_id': 43, 'end_seq_id': 49},
{'beg_seq_id': 72, 'end_seq_id': 79},
{'beg_seq_id': 89, 'end_seq_id': 93},
{'beg_seq_id': 113, 'end_seq_id': 117},
{'beg_seq_id': 139, 'end_seq_id': 141}]},
{'assignment_version': 'V1.0',
'description': 'Software generated binding site for ligand entity 3 component HEM instance E chain A',
'feature_id': 'AC3',
'name': 'ligand HEM',
'ordinal': 50,
'provenance_source': 'PDB',
'type': 'BINDING_SITE',
'feature_positions': [{'beg_seq_id': 42},
{'beg_seq_id': 43},
{'beg_seq_id': 45},
{'beg_seq_id': 46},
{'beg_seq_id': 58},
{'beg_seq_id': 61},
{'beg_seq_id': 86},
{'beg_seq_id': 87},
{'beg_seq_id': 91},
{'beg_seq_id': 93},
{'beg_seq_id': 97},
{'beg_seq_id': 98},
{'beg_seq_id': 101},
{'beg_seq_id': 136}]},
{'assignment_version': 'V1.0',
'description': 'Software generated binding site for ligand entity 3 component HEM instance G chain B',
'feature_id': 'AC4',
'name': 'ligand HEM',
'ordinal': 51,
'provenance_source': 'PDB',
'type': 'BINDING_SITE',
'feature_positions': [{'beg_seq_id': 53}]},
{'assignment_version': 'V1.0',
'description': 'Molprobity bond angle outlier in instance A model 1',
'feature_id': 'ANGLE_OUTLIER_1',
'name': 'Molprobity bond angle outlier',
'ordinal': 55,
'provenance_source': 'PDB',
'type': 'ANGLE_OUTLIER',
'feature_positions': [{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 3},
{'beg_seq_id': 3},
{'beg_seq_id': 3},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 5},
{'beg_seq_id': 5},
{'beg_seq_id': 6},
{'beg_seq_id': 6},
{'beg_seq_id': 6},
{'beg_seq_id': 6},
{'beg_seq_id': 7},
{'beg_seq_id': 7},
{'beg_seq_id': 7},
{'beg_seq_id': 7},
{'beg_seq_id': 7},
{'beg_seq_id': 7},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 10},
{'beg_seq_id': 10},
{'beg_seq_id': 11},
{'beg_seq_id': 11},
{'beg_seq_id': 11},
{'beg_seq_id': 12},
{'beg_seq_id': 12},
{'beg_seq_id': 12},
{'beg_seq_id': 12},
{'beg_seq_id': 13},
{'beg_seq_id': 13},
{'beg_seq_id': 13},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 15},
{'beg_seq_id': 15},
{'beg_seq_id': 15},
{'beg_seq_id': 15},
{'beg_seq_id': 15},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 18},
{'beg_seq_id': 18},
{'beg_seq_id': 18},
{'beg_seq_id': 19},
{'beg_seq_id': 19},
{'beg_seq_id': 19},
{'beg_seq_id': 19},
{'beg_seq_id': 19},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 22},
{'beg_seq_id': 22},
{'beg_seq_id': 22},
{'beg_seq_id': 23},
{'beg_seq_id': 23},
{'beg_seq_id': 23},
{'beg_seq_id': 23},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 25},
{'beg_seq_id': 25},
{'beg_seq_id': 26},
{'beg_seq_id': 26},
{'beg_seq_id': 26},
{'beg_seq_id': 26},
{'beg_seq_id': 26},
{'beg_seq_id': 27},
{'beg_seq_id': 27},
{'beg_seq_id': 28},
{'beg_seq_id': 28},
{'beg_seq_id': 28},
{'beg_seq_id': 29},
{'beg_seq_id': 30},
{'beg_seq_id': 30},
{'beg_seq_id': 30},
{'beg_seq_id': 31},
{'beg_seq_id': 31},
{'beg_seq_id': 31},
{'beg_seq_id': 32},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 34},
{'beg_seq_id': 34},
{'beg_seq_id': 35},
{'beg_seq_id': 36},
{'beg_seq_id': 36},
{'beg_seq_id': 38},
{'beg_seq_id': 39},
{'beg_seq_id': 39},
{'beg_seq_id': 40},
{'beg_seq_id': 41},
{'beg_seq_id': 41},
{'beg_seq_id': 42},
{'beg_seq_id': 42},
{'beg_seq_id': 42},
{'beg_seq_id': 42},
{'beg_seq_id': 42},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 44},
{'beg_seq_id': 44},
{'beg_seq_id': 45},
{'beg_seq_id': 45},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 47},
{'beg_seq_id': 47},
{'beg_seq_id': 47},
{'beg_seq_id': 47},
{'beg_seq_id': 48},
{'beg_seq_id': 48},
{'beg_seq_id': 48},
{'beg_seq_id': 49},
{'beg_seq_id': 49},
{'beg_seq_id': 49},
{'beg_seq_id': 49},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 51},
{'beg_seq_id': 51},
{'beg_seq_id': 52},
{'beg_seq_id': 52},
{'beg_seq_id': 52},
{'beg_seq_id': 52},
{'beg_seq_id': 53},
{'beg_seq_id': 54},
{'beg_seq_id': 54},
{'beg_seq_id': 54},
{'beg_seq_id': 55},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 57},
{'beg_seq_id': 57},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 59},
{'beg_seq_id': 59},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 62},
{'beg_seq_id': 62},
{'beg_seq_id': 62},
{'beg_seq_id': 62},
{'beg_seq_id': 62},
{'beg_seq_id': 63},
{'beg_seq_id': 63},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 65},
{'beg_seq_id': 65},
{'beg_seq_id': 67},
{'beg_seq_id': 67},
{'beg_seq_id': 68},
{'beg_seq_id': 69},
{'beg_seq_id': 70},
{'beg_seq_id': 70},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 73},
{'beg_seq_id': 73},
{'beg_seq_id': 73},
{'beg_seq_id': 73},
{'beg_seq_id': 73},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 76},
{'beg_seq_id': 76},
{'beg_seq_id': 76},
{'beg_seq_id': 77},
{'beg_seq_id': 77},
{'beg_seq_id': 77},
{'beg_seq_id': 77},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 79},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 82},
{'beg_seq_id': 82},
{'beg_seq_id': 82},
{'beg_seq_id': 83},
{'beg_seq_id': 83},
{'beg_seq_id': 83},
{'beg_seq_id': 83},
{'beg_seq_id': 83},
{'beg_seq_id': 83},
{'beg_seq_id': 84},
{'beg_seq_id': 84},
{'beg_seq_id': 84},
{'beg_seq_id': 84},
{'beg_seq_id': 84},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 86},
{'beg_seq_id': 86},
{'beg_seq_id': 86},
{'beg_seq_id': 87},
{'beg_seq_id': 87},
{'beg_seq_id': 88},
{'beg_seq_id': 88},
{'beg_seq_id': 88},
{'beg_seq_id': 88},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 89},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 93},
{'beg_seq_id': 94},
{'beg_seq_id': 95},
{'beg_seq_id': 96},
{'beg_seq_id': 96},
{'beg_seq_id': 96},
{'beg_seq_id': 98},
{'beg_seq_id': 98},
{'beg_seq_id': 98},
{'beg_seq_id': 98},
{'beg_seq_id': 98},
{'beg_seq_id': 99},
{'beg_seq_id': 99},
{'beg_seq_id': 99},
{'beg_seq_id': 100},
{'beg_seq_id': 101},
{'beg_seq_id': 101},
{'beg_seq_id': 103},
{'beg_seq_id': 103},
{'beg_seq_id': 103},
{'beg_seq_id': 103},
{'beg_seq_id': 104},
{'beg_seq_id': 105},
{'beg_seq_id': 105},
{'beg_seq_id': 105},
{'beg_seq_id': 106},
{'beg_seq_id': 106},
{'beg_seq_id': 106},
{'beg_seq_id': 106},
{'beg_seq_id': 108},
{'beg_seq_id': 109},
{'beg_seq_id': 109},
{'beg_seq_id': 109},
{'beg_seq_id': 110},
{'beg_seq_id': 110},
{'beg_seq_id': 110},
{'beg_seq_id': 111},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 113},
{'beg_seq_id': 113},
{'beg_seq_id': 114},
{'beg_seq_id': 114},
{'beg_seq_id': 116},
{'beg_seq_id': 116},
{'beg_seq_id': 116},
{'beg_seq_id': 117},
{'beg_seq_id': 117},
{'beg_seq_id': 117},
{'beg_seq_id': 117},
{'beg_seq_id': 117},
{'beg_seq_id': 117},
{'beg_seq_id': 118},
{'beg_seq_id': 119},
{'beg_seq_id': 120},
{'beg_seq_id': 122},
{'beg_seq_id': 125},
{'beg_seq_id': 126},
{'beg_seq_id': 126},
{'beg_seq_id': 126},
{'beg_seq_id': 126},
{'beg_seq_id': 127},
{'beg_seq_id': 127},
{'beg_seq_id': 128},
{'beg_seq_id': 128},
{'beg_seq_id': 128},
{'beg_seq_id': 128},
{'beg_seq_id': 129},
{'beg_seq_id': 131},
{'beg_seq_id': 131},
{'beg_seq_id': 131},
{'beg_seq_id': 132},
{'beg_seq_id': 132},
{'beg_seq_id': 132},
{'beg_seq_id': 134},
{'beg_seq_id': 134},
{'beg_seq_id': 134},
{'beg_seq_id': 134},
{'beg_seq_id': 135},
{'beg_seq_id': 136},
{'beg_seq_id': 136},
{'beg_seq_id': 137},
{'beg_seq_id': 137},
{'beg_seq_id': 138},
{'beg_seq_id': 138},
{'beg_seq_id': 138},
{'beg_seq_id': 139},
{'beg_seq_id': 139},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141}]},
{'assignment_version': 'V1.0',
'description': 'Molprobity bond distance outlier in instance A model 1',
'feature_id': 'BOND_OUTLIER_2',
'name': 'Molprobity bond distance outlier',
'ordinal': 56,
'provenance_source': 'PDB',
'type': 'BOND_OUTLIER',
'feature_positions': [{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 1},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 2},
{'beg_seq_id': 3},
{'beg_seq_id': 3},
{'beg_seq_id': 3},
{'beg_seq_id': 3},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 4},
{'beg_seq_id': 5},
{'beg_seq_id': 5},
{'beg_seq_id': 5},
{'beg_seq_id': 6},
{'beg_seq_id': 7},
{'beg_seq_id': 7},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 8},
{'beg_seq_id': 9},
{'beg_seq_id': 9},
{'beg_seq_id': 10},
{'beg_seq_id': 10},
{'beg_seq_id': 11},
{'beg_seq_id': 11},
{'beg_seq_id': 11},
{'beg_seq_id': 11},
{'beg_seq_id': 11},
{'beg_seq_id': 12},
{'beg_seq_id': 12},
{'beg_seq_id': 12},
{'beg_seq_id': 13},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 14},
{'beg_seq_id': 15},
{'beg_seq_id': 15},
{'beg_seq_id': 15},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 16},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 17},
{'beg_seq_id': 18},
{'beg_seq_id': 18},
{'beg_seq_id': 19},
{'beg_seq_id': 19},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 20},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 21},
{'beg_seq_id': 22},
{'beg_seq_id': 22},
{'beg_seq_id': 23},
{'beg_seq_id': 23},
{'beg_seq_id': 23},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 24},
{'beg_seq_id': 25},
{'beg_seq_id': 26},
{'beg_seq_id': 26},
{'beg_seq_id': 26},
{'beg_seq_id': 27},
{'beg_seq_id': 27},
{'beg_seq_id': 27},
{'beg_seq_id': 28},
{'beg_seq_id': 29},
{'beg_seq_id': 29},
{'beg_seq_id': 30},
{'beg_seq_id': 30},
{'beg_seq_id': 30},
{'beg_seq_id': 31},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 33},
{'beg_seq_id': 34},
{'beg_seq_id': 34},
{'beg_seq_id': 35},
{'beg_seq_id': 35},
{'beg_seq_id': 36},
{'beg_seq_id': 36},
{'beg_seq_id': 37},
{'beg_seq_id': 37},
{'beg_seq_id': 37},
{'beg_seq_id': 39},
{'beg_seq_id': 39},
{'beg_seq_id': 40},
{'beg_seq_id': 40},
{'beg_seq_id': 40},
{'beg_seq_id': 41},
{'beg_seq_id': 41},
{'beg_seq_id': 42},
{'beg_seq_id': 42},
{'beg_seq_id': 42},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 43},
{'beg_seq_id': 44},
{'beg_seq_id': 44},
{'beg_seq_id': 44},
{'beg_seq_id': 44},
{'beg_seq_id': 45},
{'beg_seq_id': 45},
{'beg_seq_id': 45},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 46},
{'beg_seq_id': 47},
{'beg_seq_id': 47},
{'beg_seq_id': 47},
{'beg_seq_id': 48},
{'beg_seq_id': 48},
{'beg_seq_id': 48},
{'beg_seq_id': 49},
{'beg_seq_id': 49},
{'beg_seq_id': 49},
{'beg_seq_id': 49},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 50},
{'beg_seq_id': 51},
{'beg_seq_id': 52},
{'beg_seq_id': 52},
{'beg_seq_id': 52},
{'beg_seq_id': 52},
{'beg_seq_id': 53},
{'beg_seq_id': 54},
{'beg_seq_id': 54},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 56},
{'beg_seq_id': 57},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 58},
{'beg_seq_id': 59},
{'beg_seq_id': 59},
{'beg_seq_id': 59},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 60},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 61},
{'beg_seq_id': 62},
{'beg_seq_id': 62},
{'beg_seq_id': 63},
{'beg_seq_id': 63},
{'beg_seq_id': 63},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 64},
{'beg_seq_id': 67},
{'beg_seq_id': 67},
{'beg_seq_id': 68},
{'beg_seq_id': 68},
{'beg_seq_id': 68},
{'beg_seq_id': 69},
{'beg_seq_id': 69},
{'beg_seq_id': 70},
{'beg_seq_id': 70},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 71},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 72},
{'beg_seq_id': 73},
{'beg_seq_id': 73},
{'beg_seq_id': 73},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 74},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 75},
{'beg_seq_id': 76},
{'beg_seq_id': 76},
{'beg_seq_id': 76},
{'beg_seq_id': 76},
{'beg_seq_id': 76},
{'beg_seq_id': 77},
{'beg_seq_id': 77},
{'beg_seq_id': 77},
{'beg_seq_id': 77},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 78},
{'beg_seq_id': 79},
{'beg_seq_id': 79},
{'beg_seq_id': 79},
{'beg_seq_id': 80},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 81},
{'beg_seq_id': 83},
{'beg_seq_id': 84},
{'beg_seq_id': 84},
{'beg_seq_id': 84},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 85},
{'beg_seq_id': 86},
{'beg_seq_id': 86},
{'beg_seq_id': 87},
{'beg_seq_id': 87},
{'beg_seq_id': 87},
{'beg_seq_id': 88},
{'beg_seq_id': 88},
{'beg_seq_id': 88},
{'beg_seq_id': 89},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 90},
{'beg_seq_id': 91},
{'beg_seq_id': 91},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 92},
{'beg_seq_id': 93},
{'beg_seq_id': 93},
{'beg_seq_id': 94},
{'beg_seq_id': 95},
{'beg_seq_id': 96},
{'beg_seq_id': 96},
{'beg_seq_id': 96},
{'beg_seq_id': 97},
{'beg_seq_id': 98},
{'beg_seq_id': 98},
{'beg_seq_id': 99},
{'beg_seq_id': 99},
{'beg_seq_id': 99},
{'beg_seq_id': 99},
{'beg_seq_id': 99},
{'beg_seq_id': 104},
{'beg_seq_id': 105},
{'beg_seq_id': 105},
{'beg_seq_id': 105},
{'beg_seq_id': 106},
{'beg_seq_id': 106},
{'beg_seq_id': 106},
{'beg_seq_id': 107},
{'beg_seq_id': 107},
{'beg_seq_id': 108},
{'beg_seq_id': 109},
{'beg_seq_id': 109},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 112},
{'beg_seq_id': 113},
{'beg_seq_id': 113},
{'beg_seq_id': 114},
{'beg_seq_id': 114},
{'beg_seq_id': 114},
{'beg_seq_id': 114},
{'beg_seq_id': 115},
{'beg_seq_id': 115},
{'beg_seq_id': 116},
{'beg_seq_id': 117},
{'beg_seq_id': 117},
{'beg_seq_id': 119},
{'beg_seq_id': 120},
{'beg_seq_id': 120},
{'beg_seq_id': 121},
{'beg_seq_id': 122},
{'beg_seq_id': 122},
{'beg_seq_id': 123},
{'beg_seq_id': 127},
{'beg_seq_id': 127},
{'beg_seq_id': 127},
{'beg_seq_id': 128},
{'beg_seq_id': 128},
{'beg_seq_id': 128},
{'beg_seq_id': 129},
{'beg_seq_id': 129},
{'beg_seq_id': 131},
{'beg_seq_id': 131},
{'beg_seq_id': 132},
{'beg_seq_id': 132},
{'beg_seq_id': 134},
{'beg_seq_id': 134},
{'beg_seq_id': 135},
{'beg_seq_id': 137},
{'beg_seq_id': 137},
{'beg_seq_id': 137},
{'beg_seq_id': 137},
{'beg_seq_id': 137},
{'beg_seq_id': 138},
{'beg_seq_id': 138},
{'beg_seq_id': 139},
{'beg_seq_id': 139},
{'beg_seq_id': 139},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 140},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141},
{'beg_seq_id': 141}]},
{'assignment_version': 'V1.0',
'description': 'Molprobity Ramachandran outlier in instance A model 1',
'feature_id': 'RAMACHANDRAN_OUTLIER_3',
'name': 'Molprobity Ramachandran outlier',
'ordinal': 57,
'provenance_source': 'PDB',
'type': 'RAMACHANDRAN_OUTLIER',
'feature_positions': [{'beg_seq_id': 3},
{'beg_seq_id': 16},
{'beg_seq_id': 21}]},
{'assignment_version': 'V1.0',
'description': 'Molprobity rotamer outlier in instance A model 1',
'feature_id': 'ROTAMER_OUTLIER_4',
'name': 'Molprobity rotamer outlier',
'ordinal': 58,
'provenance_source': 'PDB',
'type': 'ROTAMER_OUTLIER',
'feature_positions': [{'beg_seq_id': 2},
{'beg_seq_id': 4},
{'beg_seq_id': 45},
{'beg_seq_id': 52},
{'beg_seq_id': 75},
{'beg_seq_id': 84},
{'beg_seq_id': 95},
{'beg_seq_id': 138}]},
{'assignment_version': 'V1.0',
'description': 'STEREO_OUTLIER in instance A model 1',
'feature_id': 'STEREO_OUTLIER_5',
'name': 'STEREO_OUTLIER',
'ordinal': 59,
'provenance_source': 'PDB',
'type': 'STEREO_OUTLIER',
'feature_positions': [{'beg_seq_id': 137}]},
{'description': 'The Accessible Surface Area considering this polymer entity instance by itself (unbound).',
'name': 'Unbound ASA',
'ordinal': 60,
'provenance_source': 'null-null',
'type': 'ASA',
'feature_positions': [{'beg_seq_id': 1,
'end_seq_id': 141,
'values': [163.21193608797677,
18.05226368835482,
42.73879697266144,
107.38936955373997,
66.50730410483254,
13.991379449108642,
48.67727291088148,
85.28624205006685,
34.520959847459366,
1.3437094434228118,
92.863304857998,
65.47356555544295,
2.4012047164588566,
21.923797855183835,
53.0742572985793,
120.18688975909083,
0.7884492781826147,
43.17696746642822,
110.12019771123582,
81.17388093023601,
7.552498066654208,
18.85531760246544,
113.94073377620057,
14.61770997999541,
2.340662456112997,
13.526881152267718,
39.64487610263428,
0.2687418886845624,
3.4936445528993114,
47.89055402193245,
87.70392630494936,
9.092980537616087,
12.08677005331391,
135.11954691588275,
69.80058918801944,
57.82767547901989,
74.1190475823852,
94.89898526921169,
2.517197343777638,
87.87390908697569,
99.65368019290668,
72.37646394274378,
31.11039282732865,
109.87626058150693,
125.39761701574386,
47.02814411286198,
74.27110695000425,
53.52575065847143,
60.67111088359101,
159.16967520998105,
34.21547012118724,
12.508562796259,
86.24288842901149,
78.15149915147933,
8.86848232659056,
127.58639603342641,
34.84356120059708,
48.02758024038556,
0.0,
139.02760775217652,
139.50191535791902,
27.671383083949387,
1.4780803877650932,
80.69898396171169,
21.344669125598724,
11.690272157778464,
56.79320179694933,
54.39928054889856,
0.2687418886845624,
12.353238685554334,
62.651300715445814,
92.26515943976219,
11.307510561961573,
114.67969920017796,
65.80122989930118,
9.0673350884663,
85.31542870245575,
103.18286998172576,
27.417676857704905,
4.416464775424207,
46.9556009685794,
78.64429817158087,
53.89481365368161,
1.4151354373577678,
61.48191699173364,
76.57396599339808,
34.59920184081555,
7.68458695809292,
105.82181535056358,
158.3769646755231,
88.65497687905943,
161.60157203002842,
23.78365714858377,
72.80740751677023,
63.2790122904631,
70.60291425263443,
18.667783370602052,
23.703777757136542,
146.79132263275392,
42.72996030084542,
38.16134819320786,
15.623877912257104,
96.23684506711352,
3.9836073431531753,
10.212191770013371,
13.885696272241882,
53.22958769246928,
0.1343709443422812,
5.374837773691248,
38.00745162220499,
64.76393381039064,
65.20523835698864,
4.702983051979842,
113.7395461826647,
84.57884586234685,
46.23905897632628,
44.632259997721235,
62.02522151197134,
117.39509334128527,
64.49508887746705,
20.29001259568446,
82.72431722336152,
59.32964328068581,
2.2659679491812454,
2.418676998161062,
70.20040837530826,
79.8176717179631,
1.8822412561011748,
10.400201010685985,
58.28403453761389,
29.70257353094256,
10.346562714355652,
18.462073140282055,
84.21967511408428,
18.140077486207964,
21.096238261738147,
34.88131057792261,
70.15298163597261,
75.58824103285183,
56.74619728766633,
281.2892740261974]}]}],
'rcsb_polymer_instance_feature_summary': [{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'BEND'},
{'count': 2,
'coverage': 0.10638,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'BINDING_SITE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'C-MANNOSYLATION_SITE'},
{'count': 1,
'coverage': 1.0,
'maximum_length': 141,
'maximum_value': 0.0,
'minimum_length': 141,
'minimum_value': 0.0,
'type': 'CATH'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'CIS-PEPTIDE'},
{'count': 1,
'coverage': 1.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'ECOD'},
{'count': 8,
'coverage': 0.78014,
'maximum_length': 21,
'maximum_value': 0.0,
'minimum_length': 2,
'minimum_value': 0.0,
'type': 'HELIX_P'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'HELX_LH_PP_P'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'HELX_RH_3T_P'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'HELX_RH_AL_P'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'HELX_RH_PI_P'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_CONTACT_PROBABILITY'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_DISTANCE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_ENERGY'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_IPTM'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_NORMALIZED_SCORE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_OTHER'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_PAE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_PLDDT'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_PLDDT_ALL-ATOM'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_PLDDT_ALL-ATOM_[0,1]'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_PLDDT_[0,1]'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_PTM'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MA_QA_METRIC_LOCAL_TYPE_ZSCORE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'MEMBRANE_SEGMENT'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'N-GLYCOSYLATION_SITE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'O-GLYCOSYLATION_SITE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'S-GLYCOSYLATION_SITE'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SABDAB_ANTIBODY_HEAVY_CHAIN_SUBCLASS'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SABDAB_ANTIBODY_LIGHT_CHAIN_SUBCLASS'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SABDAB_ANTIBODY_LIGHT_CHAIN_TYPE'},
{'count': 1,
'coverage': 1.0,
'maximum_length': 141,
'maximum_value': 0.0,
'minimum_length': 141,
'minimum_value': 0.0,
'type': 'SCOP'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SCOP2B_SUPERFAMILY'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SCOP2_FAMILY'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SCOP2_SUPERFAMILY'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'SHEET'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'STRN'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'TURN_TY1_P'},
{'count': 1,
'coverage': 0.21986,
'maximum_length': 31,
'maximum_value': 0.0,
'minimum_length': 31,
'minimum_value': 0.0,
'type': 'UNASSIGNED_SEC_STRUCT'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'UNOBSERVED_ATOM_XYZ'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'UNOBSERVED_RESIDUE_XYZ'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'ZERO_OCCUPANCY_ATOM_XYZ'},
{'count': 0,
'coverage': 0.0,
'maximum_length': 0,
'maximum_value': 0.0,
'minimum_length': 0,
'minimum_value': 0.0,
'type': 'ZERO_OCCUPANCY_RESIDUE_XYZ'},
{'count': 444, 'coverage': 3.14894, 'type': 'ANGLE_OUTLIER'},
{'count': 372, 'coverage': 2.6383, 'type': 'BOND_OUTLIER'},
{'count': 0, 'coverage': 0.0, 'type': 'MOGUL_ANGLE_OUTLIER'},
{'count': 0, 'coverage': 0.0, 'type': 'MOGUL_BOND_OUTLIER'},
{'count': 3, 'coverage': 0.02128, 'type': 'RAMACHANDRAN_OUTLIER'},
{'count': 8, 'coverage': 0.05674, 'type': 'ROTAMER_OUTLIER'},
{'count': 0, 'coverage': 0.0, 'type': 'RSCC_OUTLIER'},
{'count': 0, 'coverage': 0.0, 'type': 'RSRZ_OUTLIER'},
{'count': 1, 'coverage': 0.00709, 'type': 'STEREO_OUTLIER'}],
'rcsb_polymer_struct_conn': [{'connect_type': 'metal coordination',
'dist_value': 2.143,
'id': 'metalc1',
'ordinal_id': 1,
'connect_target': {'auth_seq_id': '87',
'label_asym_id': 'A',
'label_atom_id': 'NE2',
'label_comp_id': 'HIS',
'label_seq_id': 87,
'symmetry': '1_555'},
'connect_partner': {'label_asym_id': 'E',
'label_atom_id': 'FE',
'label_comp_id': 'HEM',
'symmetry': '1_555'}}],
'rcsb_id': '4HHB.A',
'rcsb_latest_revision': {'major_revision': 4, 'minor_revision': 1}}
BLAST URLAPI¶
http://www.ncbi.nlm.nih.gov/BLAST/Doc/urlapi.html
A URLAPI request looks like this:
http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=<command>&{<name>=<value>}
where CMD can be
- PUT add a job to the queue
- GET get formated results from a job
BLAST PUT¶
Some common attributes to PUT:
- DATABASE - what database to search, mandatory
- PROGRAM - what program to use (blastn, blastp, blastx, tblastn, tblastx)
- QUERY - Accession(s), gi(s), or FASTA sequence(s), mandatory
- MATRIX_NAME - matrix to use (default BLOSUM62)
%%html
<div id="webblast" style="width: 500px"></div>
<script>
var divid = '#webblast';
jQuery(divid).asker({
id: divid,
question: "When making a BLAST request with a long sequence, which HTTP request type should you use?",
answers: ['GET','POST','REQUEST','Does not matter'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
response = requests.post('https://www.ncbi.nlm.nih.gov/blast/Blast.cgi',\
data={'CMD': 'PUT', 'DATABASE': 'nr', 'PROGRAM':'blastp', \
'QUERY': 'SQETFSDLWKLLPEN'})
print(response.text)
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" lang="en">
<head>
<meta http-equiv="Content-Type" content="text/html; charset=UTF-8" />
<meta name="jig" content="ncbitoggler ncbiautocomplete"/>
<meta name="ncbi_app" content="static" />
<meta name="ncbi_pdid" content="blastformatreq" />
<meta name="ncbi_stat" content="false" />
<meta name="ncbi_sessionid" content="21915490536CA8C1_0000SID" />
<meta name="ncbi_phid" content="21915490536CA8C10000000000000001" />
<title>NCBI Blast</title>
<meta http-equiv="Pragma" content="no-cache">
<link rel="stylesheet" type="text/css" href="css/uswds.min.css" media="screen" />
<link rel="stylesheet" type="text/css" href="https://www.ncbi.nlm.nih.gov/style-guide/static/nwds/css/nwds.css"/>
<link rel="stylesheet" href="css/headerNew.css?v=1"/>
<link rel="stylesheet" href="https://use.fontawesome.com/releases/v5.5.0/css/all.css" crossorigin="anonymous"> <!-- Font Awesome icons -->
<link rel="stylesheet" type="text/css" href="css/footerNew.css?v=1" media="screen" />
<link rel="stylesheet" type="text/css" href="css/main.css" media="screen" />
<link rel="stylesheet" type="text/css" href="css/common.css" media="screen" />
<link rel="stylesheet" type="text/css" href="css/blastReq.css" media="screen" />
<!--[if IE]>
<link rel="stylesheet" type="text/css" href="css/blastReqIE.css" media="screen" />
<![endif]-->
<link rel="stylesheet" type="text/css" href="css/print.css" media="print" />
<!--[if lte IE 6]>
<link rel="stylesheet" type="text/css" href="css/ie6_or_less.css" />
<![endif]-->
<script type="text/javascript" src="/core/jig/1.15.2/js/jig.min.js "></script>
<script type="text/javascript" src="js/utils.js"></script>
<script type="text/javascript" src="js/blast.js"></script>
<script type="text/javascript" src="js/format.js"></script>
</head>
<body id="type-a">
<div id="wrap">
<script>var useOfficialGovtHeader = true;</script>
<section class="usa-banner">
<div class="usa-accordion">
<header class="usa-banner-header">
<div class="usa-grid usa-banner-inner">
<img src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/favicons/favicon-57.png" alt="U.S. flag">
<p>An official website of the United States government</p>
<button class="usa-accordion-button usa-banner-button" aria-expanded="false" aria-controls="gov-banner-top">
<span class="usa-banner-button-text">Here's how you know</span>
</button>
</div>
</header>
<div class="usa-banner-content usa-grid usa-accordion-content" id="gov-banner-top" aria-hidden="true">
<div class="usa-banner-guidance-gov usa-width-one-half">
<img class="usa-banner-icon usa-media_block-img" src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/icon-dot-gov.svg" alt="Dot gov">
<div class="usa-media_block-body">
<p>
<strong>The .gov means itâs official.</strong>
<br>
Federal government websites often end in .gov or .mil. Before
sharing sensitive information, make sure youâre on a federal
government site.
</p>
</div>
</div>
<div class="usa-banner-guidance-ssl usa-width-one-half">
<img class="usa-banner-icon usa-media_block-img" src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/icon-https.svg" alt="Https">
<div class="usa-media_block-body">
<p>
<strong>The site is secure.</strong>
<br>
The <strong>https://</strong> ensures that you are connecting to the
official website and that any information you provide is encrypted
and transmitted securely.
</p>
</div>
</div>
</div>
</div>
</section>
<ul class="msg" id="msgServ"><li class="warning">
<p class="warning">
This URL will be replaced with <a href="https://blast.ncbi.nlm.nih.gov" title="BLAST">blast.ncbi.nlm.nih.gov</a> on 12/1/2014.
Please change your bookmark now.
</p>
</li></ul>
<header class="ncbi-header" role="banner" data-section="Header">
<a class="usa-skipnav" href="#mainCont">Skip to main page content</a>
<div class="usa-grid">
<div class="usa-width-one-whole">
<div class="ncbi-header__logo">
<a href="https://www.ncbi.nlm.nih.gov/" class="logo" aria-label="NCBI Logo" data-ga-action="click_image" data-ga-label="NIH NLM Logo">
<img src="https://www.ncbi.nlm.nih.gov/coreutils/nwds/img/logos/AgencyLogo.svg" alt="NIH NLM Logo">
</a>
</div>
<div class="ncbi-header__account">
<a id="account_login" href="https://www.ncbi.nlm.nih.gov/account/?back_url=https%3A%2F%2Fwww%2Encbi%2Enlm%2Enih%2Egov%2Fblast%2FBlast%2Ecgi%3FCMD%3DPUT%26DATABASE%3Dnr%26ENTREZ%5FQUERY%3D%26FULL%5FDBNAME%3Dnr%26JOB%5FTITLE%3DProtein%2BSequence%2B%26MYNCBI%5FUSER%3D4643079524%26MYNCBI%5FUSER%3D4643079524%26ORG%5FDBS%3Dgiless%5Fdbvers5%26PROGRAM%3Dblastp%26QUERY%5FINFO%3DProtein%2BSequence%2B%26QUERY%5FLENGTH%3D15%26RID%3DKDJMCZ64013%26RTOE%3D21%26USER%5FTYPE%3D2%26USER%5FTYPE%3D2%26WORD%5FSIZE%3D5" class="usa-button header-button">Log in</a>
<button id="account_info" class="header-button" aria-controls="account_popup">
<span class="fa fa-user" aria-hidden="true"></span>
<span class="username desktop-only" aria-hidden="true" id="uname_short"></span>
<span class="sr-only">Show account info</span>
</button>
</div>
<div class="ncbi-popup-anchor">
<div class="ncbi-popup account-popup" id="account_popup" aria-hidden="true" role="dialog" aria-labelledby="account-popup-header">
<div class="ncbi-popup-head">
<button class="ncbi-close-button"><span class="fa fa-window-close"></span><span class="usa-sr-only">Close</span></button>
<h4>Account</h4>
</div>
<div class="account-user-info">
Logged in as:<br>
<b><span class="username" id="uname_long">username</span></b>
</div>
<div class="account-links">
<ul class="usa-unstyled-list">
<li><a id="account_myncbi" href="https://www.ncbi.nlm.nih.gov/myncbi/">Dashboard</a> <span class="ncbi-text-small-light">(My NCBI)</span></li>
<li><a id="account_pubs" href="https://www.ncbi.nlm.nih.gov/myncbi/collections/bibliography/">Publications</a> <span class="ncbi-text-small-light">(My Bibliography)</span></li>
<li><a id="account_settings" href="https://www.ncbi.nlm.nih.gov/account/settings/">Account settings</a></li>
<li><a id="account_logout" href="https://www.ncbi.nlm.nih.gov/account/signout/?back_url=https%3A%2F%2Fwww%2Encbi%2Enlm%2Enih%2Egov%2Fblast%2FBlast%2Ecgi%3FCMD%3DPUT%26DATABASE%3Dnr%26ENTREZ%5FQUERY%3D%26FULL%5FDBNAME%3Dnr%26JOB%5FTITLE%3DProtein%2BSequence%2B%26MYNCBI%5FUSER%3D4643079524%26MYNCBI%5FUSER%3D4643079524%26ORG%5FDBS%3Dgiless%5Fdbvers5%26PROGRAM%3Dblastp%26QUERY%5FINFO%3DProtein%2BSequence%2B%26QUERY%5FLENGTH%3D15%26RID%3DKDJMCZ64013%26RTOE%3D21%26USER%5FTYPE%3D2%26USER%5FTYPE%3D2%26WORD%5FSIZE%3D5">Log out</a></li>
</ul>
</div>
</div>
</div>
</div>
</div>
</header>
<div role="navigation" aria-label="access keys">
<a id="nws_header_accesskey_0" href="https://www.ncbi.nlm.nih.gov/guide/browsers/#ncbi_accesskeys" class="usa-sr-only" accesskey="0" tabindex="-1">Access keys</a>
<a id="nws_header_accesskey_1" href="https://www.ncbi.nlm.nih.gov" class="usa-sr-only" accesskey="1" tabindex="-1">NCBI Homepage</a>
<a id="nws_header_accesskey_2" href="/myncbi/" class="set-base-url usa-sr-only" accesskey="2" tabindex="-1">MyNCBI Homepage</a>
<a id="nws_header_accesskey_3" href="#maincontent" class="usa-sr-only" accesskey="3" tabindex="-1">Main Content</a>
<a id="nws_header_accesskey_4" href="#" class="usa-sr-only" accesskey="4" tabindex="-1">Main Navigation</a>
</div>
<nav class="ncbi-topnav" id="navcontent">
<div class="usa-grid">
<a class="ncbi-topnav-root" href="Blast.cgi">BLAST <sup>®</sup></a> <span id="brc"><span class="brcrmbsign">»</span> blastp suite</span>
<ul class="rf ncbi-topnav-list" id="topnav-list">
<li class="first "><a href="Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome" title="BLAST Home">Home</a></li>
<li class="recent "><a href="Blast.cgi?CMD=GetSaved&RECENT_RESULTS=on" title="Unexpired BLAST jobs">Recent Results</a></li>
<li class="saved "><a href="Blast.cgi?CMD=GetSaved" title="Saved sets of BLAST search parameters">Saved Strategies</a></li>
<li class= "last documentation "> <a href="../doc/blast-help/" title="BLAST documentation">Help</a></li>
</ul>
</div>
</nav>
<div id="banner_ad" class="static">
<div>
<div><i class="fas fa-user-friends"></i><span>Check out the ClusteredNR database on BLAST+</span></div>
</div>
<a class="sbt usa-button-primary" href="https://blast.ncbi.nlm.nih.gov/doc/blast-news/2023-BLAST-News.html#thu-24-aug-2023">Learn more</a>
<a class="sbt usa-button-primary" href="https://support.nlm.nih.gov/support/create-case/">Give us feedback</a>
<button class="close" id="closeAd" aria-controls="banner_ad"><i class="fas fa-times"></i></button>
</div>
<div id="content-wrap">
<div class="pageTitle">
Format Request
<span id="frmRequestPrTr"></span>
</div>
<!-- Do errors this way -->
<!--<ul class="msg"><li class=""><p></p></li></ul>-->
<ul id="msgR" class="msg"><li class=""></li></ul>
<div id="content">
<form action="Blast.cgi" enctype="application/x-www-form-urlencoded" method="post" name="FormatForm" id="FormatForm">
<script language="JavaScript">
<!--
//document.images['BlastHeaderGif'].src = 'html/head_formating.gif';
// -->
</script>
<!--
<p class='info'>
<strong>Job submitted.</strong>
We estimate that results will be ready in 16 seconds or less.
</p>
-->
<div class="fbtn">
<!--
<a href="javascript:document.forms[0].submit();">
<img align="middle" alt="Format button" border="0" src="FormatPage_files/format_but.gif">
</a>
-->
</div>
<dl class="summary query title db">
<dd>
</dd>
<!-- <span class=" query title db">-->
<dt class="hidden query">Query</dt><dd class="hidden query">Protein Sequence</dd>
<dt class="hidden db">Database</dt><dd class="hidden db">nr</dd>
<dt class="hidden title">Job title</dt><dd class="hidden title">Protein Sequence</dd>
<dt class="hidden entrez">Entrez Query</dt><dd class="hidden entrez"><span class="note entrez">Note: Your search is limited to records matching this Entrez query</span></dd>
<!-- </span> -->
<dt><label for="rid">Request ID</label></dt><dd><input name="RID" size="50" type="text" value="KDJMCZ64013" id="rid" />
<input type="submit" value="View report" name="ViewReport" class="button" />
<!-- <img border="0" id="viewRpButton" src="images/veiwRpButton.jpg" class="viewReport" alt="View report" mouseovImg="images/veiwRpButtonOver.jpg" mouseoutImg="images/veiwRpButton.jpg" mousedownImg="images/veiwRpButtonDown.jpg" mouseupImg="images/veiwRpButtonOver.jpg" />-->
<input type="checkbox" name="NEWWINRES" form="FormatForm" winType="const" id="nw" class="newwin" />
<label for="nw">Show results in a new window</label>
</dd>
<dt>Format<br/>
<!--<a class='help' href="#">[Help]</a></dt> -->
<dd>
<table id="filterResults" class="options blastp ">
<tr class="paramSet xgl">
<td class="hd"><label for="FORMAT_OBJECT">Show</label></td>
<td>
<div class="fi">
<select id="FORMAT_OBJECT" class="reset" name="FORMAT_OBJECT" defVal="Alignment">
<option value="Alignment" >Alignment</option>
<option value="PSSM_Scoremat" >PssmWithParameters</option>
<option value="Bioseq" >Bioseq</option>
</select>
<label for="FORMAT_TYPE">as</label>
<select name="FORMAT_TYPE" id="FORMAT_TYPE" class="reset" defVal="HTML">
<option value="HTML" >HTML</option>
<option value="Text" >Plain text</option>
<option value="ASN.1" >ASN.1</option>
<option value="XML" >XML</option>
<option value="XML2" >XML2</option>
<option value="JSON2" >JSON2</option>
<option value="XML2_S" >XML2_S</option>
<option value="JSON2_S" >JSON2_S</option>
<option value="SAM_SQ" >SAM_SQ</option>
<option value="SAM" >SAM</option>
</select>
<input name="PSSM_FORMAT_TYPE" value="Text" size="3" id="pssmFormat" type="text" class="hidden dispType" />
<input name="BIOSEQ_FORMAT_TYPE" value="ASN.1" size="3" id="bioseqFormat" type="text" class="hidden dispType" />
<input name="PSSM_SC_FORMAT_TYPE" value="ASN.1" size="3" id="pssmScFormat" type="text" class="hidden dispType" />
<a class="resetAll" id="resetAll" >Reset form to defaults</a>
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Alignments object formatting help" id="formatHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<p class="helpbox ui-ncbitoggler-slave" id="hlp1">
These options control formatting of alignments in results pages. The
default is HTML, but other formats (including plain text) are available.
PSSM and PssmWithParameters are representations of Position Specific Scoring Matrices and are only available for PSI-BLAST.
The Advanced view option allows the database descriptions to be sorted by various indices in a table.
</p>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="odd paramSet">
<td class="hd"><label for="ALIGNMENT_VIEW">Alignment View</label></td>
<td>
<div class="fi">
<select name="ALIGNMENT_VIEW" id="ALIGNMENT_VIEW" defVal="Pairwise" class="reset">
<option value="Pairwise" >Pairwise</option>
<option value="PairwiseWithIdentities" >Pairwise with dots for identities</option>
<option value="QueryAnchored" >Query-anchored with dots for identities</option>
<option value="QueryAnchoredNoIdentities" >Query-anchored with letters for identities</option>
<option value="FlatQueryAnchored" >Flat query-anchored with dots for identities</option>
<option value="FlatQueryAnchoredNoIdentities" >Flat query-anchored with letters for identities</option>
<option value="Tabular" >Hit Table</option>
</select>
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Alignments view options help" id="alnViewHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<p class="helpbox ui-ncbitoggler-slave" id="hlp2">
Choose how to view alignments.
The default "pairwise" view shows how each subject sequence aligns
individually to the query sequence. The "query-anchored" view shows how
all subject sequences align to the query sequence. For each view type,
you can choose to show "identities" (matching residues) as letters or
dots.
<a href="https://blast.ncbi.nlm.nih.gov/doc/blast-topics/resultformatoptions.html#alignment-view" target="helpWin" title="Additional alignments view options help">more...</a>
</p>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="paramSet">
<td class="hd"><label>Display</label></td>
<td class="cb">
<div class="fi">
<input name="SHOW_OVERVIEW" id="SHOW_OVERVIEW" type="checkbox" class="cb reset" defVal="checked" checked="checked" />
<label class="rb" for="SHOW_OVERVIEW">Graphical Overview</label>
<span id="shl" >
<input name="SHOW_LINKOUT" id="SHOW_LINKOUT" type="checkbox" class="cb reset" defVal="checked" checked="checked" />
<label class="rb" for="SHOW_LINKOUT">Linkout</label>
</span>
<span id="gts" >
<input name="GET_SEQUENCE" id="GET_SEQUENCE" type="checkbox" class="cb reset" defVal="checked" checked="checked" />
<label class="rb" for="GET_SEQUENCE">Sequence Retrieval</label>
</span>
<input name="NCBI_GI" id="NCBI_GI" type="checkbox" class="cb reset hidden" defVal="unchecked" />
<label class="rb hidden" for="NCBI_GI">NCBI-gi</label>
<span id="scf" >
<input name="SHOW_CDS_FEATURE" id="SHOW_CDS_FEATURE" type="checkbox" class="cb reset blastn" defVal="unchecked" />
<label for="SHOW_CDS_FEATURE" class="blastn">CDS feature</label>
</span>
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Alignments display options help" id="displayHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<ul class="helpbox ui-ncbitoggler-slave" id="hlp3">
<li>Graphical Overview: Graphical Overview: Show graph of similar sequence regions aligned to query.
<a href="https://blast.ncbi.nlm.nih.gov/doc/blast-topics/resultformatoptions.html#graphical-overview" target="helpWin" title="Graphical Overview help">more...</a>
</li>
<li>NCBI-gi: Show NCBI gi identifiers.
</li>
<li>CDS feature: Show annotated coding region and translation.
<a href="https://blast.ncbi.nlm.nih.gov/doc/blast-topics/resultformatoptions.html#cds-feature" title="CDS feature help" target="helpWin" >more...</a>
</li></ul>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="paramSet odd xgl">
<td class="hd"><label>Masking</label></td>
<td>
<div class="fi">
<label for="MASK_CHAR"> Character: </label>
<select name="MASK_CHAR" id="MASK_CHAR" class="reset" defVal="2">
<option value="0" >X for protein, n for nucleotide</option>
<option value="2" selected="selected" >Lower Case</option>
</select>
<label for="MASK_COLOR"> Color:</label>
<select name="MASK_COLOR" id="MASK_COLOR" class="reset" defVal="1">
<option value="0" >Black
</option>
<option value="1" selected="selected" >Grey
</option>
<option value="2" >Red
</option>
</select>
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Alignments masking help" id="maskingHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<ul class="helpbox ui-ncbitoggler-slave" id="hlp4">
<li>Masking Character: Display masked (filtered) sequence regions as lower-case or as specific letters (N for nucleotide, P for protein).
</li>
<li>Masking Color: Display masked sequence regions in the given color.</li>
</ul>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr id="lr" class="paramSet xgl">
<td class="hd"><label>Limit results</label></td>
<td>
<div class="fi">
<label for="FRM_DESCRIPTIONS">Descriptions:</label>
<select name="DESCRIPTIONS" id="FRM_DESCRIPTIONS" class="reset" defVal="100">
<option value="0" >0</option>
<option value="10" >10</option>
<option value="50" >50</option>
<option value="100" selected="selected" >100</option>
<option value="250" >250</option>
<option value="500" >500</option>
<option value="1000" >1000</option>
<option value="5000" >5000</option>
<option value="10000" >10000</option>
<option value="20000" >20000</option>
</select>
<label for="FRM_NUM_OVERVIEW">Graphical overview:</label>
<select name="NUM_OVERVIEW" id="FRM_NUM_OVERVIEW" class="reset" defVal="100">
<option value="0" >0</option>
<option value="10" >10</option>
<option value="50" >50</option>
<option value="100" selected="selected" >100</option>
<option value="250" >250</option>
<option value="500" >500</option>
<option value="1000" >1000</option>
</select>
<span id="frmAln">
<label for="FRM_ALIGNMENTS">Alignments:</label>
<select name="ALIGNMENTS" id="FRM_ALIGNMENTS" class="reset" defVal="100">
<option value="0" >0</option>
<option value="10" >10</option>
<option value="50" >50</option>
<option value="100" selected="selected" >100</option>
<option value="250" >250</option>
<option value="500" >500</option>
<option value="1000" >1000</option>
<option value="5000" >5000</option>
<option value="10000" >10000</option>
<option value="20000" >20000</option>
</select>
</span>
<label for="FRM_LINE_LENGTH">Line length:</label>
<select name="LINE_LENGTH" id="FRM_LINE_LENGTH" class="reset" defVal="60">
<option value="60" >60</option>
<option value="90" >90</option>
<option value="120" >120</option>
<option value="150" >150</option>
</select>
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Limit number of descriptions/alignments help" id="numHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<ul class="helpbox ui-ncbitoggler-slave" id="hlp5">
<li>Descriptions: Show short descriptions for up to the given number of sequences.</li>
<li>Alignments: Show alignments for up to the given number of sequences, in order of statistical significance.</li>
<li>Line lenghth: Number of letters to show on one line in an alignment.</li>
</ul>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="paramSet odd xgl ">
<td class="hd"></td>
<td>
<div class="">
<label for="qorganism">Organism</label>
<span class="instr">Type common name, binomial, taxid, or group name. Only 20 top taxa will be shown.</span><br/>
<input name="FORMAT_ORGANISM" size="55" type="text" id="qorganism" value="" data-jigconfig="dictionary:'blast_nt_nucl_sg',isCrossDomain:false" autocomplete="off" data-jig="ncbiautocomplete" class="reset">
<input type="hidden" value = "1" name="FORMAT_NUM_ORG" id="numOrg" />
<input type="checkbox" name="FORMAT_ORG_EXCLUDE" class="oExclR cb" id="orgExcl"/>
<label for="orgExcl" class="right">exclude</label>
<a href="#" title="Add organism" class="addOrg" id="addOrg"><img border="0" src="css/images/addOrg.jpg" id="addOrgIm" alt="Add organism" mouseovImg="css/images/addOrgOver.jpg" mouseoutImg="css/images/addOrg.jpg" mousedownImg="css/images/addOrgDown.jpg" mouseupImg="css/images/addOrgOver.jpg" /></a>
<div id="orgs">
</div>
<div class="fi">
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Limit results by organism help" id="organismHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<p class="helpbox ui-ncbitoggler-slave" id="hlp6">
Show only sequences from the given organism.
</p>
</div><!-- ARIA -->
</div>
</div>
</td>
</tr>
<tr class="paramSet xgl hidden">
<td class="hd"></td>
<td>
<div class="fi">
<label for="FORMAT_EQ_TEXT">Entrez query:</label>
<input name="FORMAT_EQ_TEXT" id="FORMAT_EQ_TEXT" size="60" type="text" value="" class="reset" />
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Limit results by Entrez query help" id="entrezHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<p class="helpbox ui-ncbitoggler-slave" id="hlp7">
Show only those sequences that match the given Entrez query.
<a href="https://blast.ncbi.nlm.nih.gov/doc/blast-topics/resultformatoptions.html#limit-results-by-entrez-query" target="helpWin" title="Additional limit results by Entrez query help" target="helpWin">more...</a>
</p>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="paramSet odd xgl">
<td class="hd"></td>
<td>
<div class="fi">
<label for="EXPECT_LOW">Expect Min:</label> <input name="EXPECT_LOW" id="EXPECT_LOW" size="10" type="text" value="" class="reset"/>
<label for="EXPECT_HIGH">Expect Max:</label> <input name="EXPECT_HIGH" id="EXPECT_HIGH" size="10" type="text" value="" class="reset" />
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Limit results by expect value range help" id="expectHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<p class="helpbox ui-ncbitoggler-slave" id="hlp8">
Show only sequences with expect values in the given range.
<a href="https://blast.ncbi.nlm.nih.gov/doc/blast-topics/resultformatoptions.html#expect-value-range" target="helpWin" title="Additional limit results by expect value range help">more...</a>
</p>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="paramSet xgl">
<td class="hd"></td>
<td>
<div class="fi">
<label for="PERC_IDENT_LOW">Percent Identity Min:</label> <input name="PERC_IDENT_LOW" id="PERC_IDENT_LOW" size="10" type="text" value="" class="reset"/>
<label for="PERC_IDENT_HIGH">Percent Identity Max:</label> <input name="PERC_IDENT_HIGH" id="PERC_IDENT_HIGH" size="10" type="text" value="" class="reset" />
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="Limit results by percent identity range help" id="percIdentHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<p class="helpbox ui-ncbitoggler-slave" id="hlp10">
Show only sequences with percent identity values in the given range.
</p>
</div><!-- ARIA -->
</div>
</td>
</tr>
<tr class="psiBlast odd paramSet xgl ">
<td class="hd"><label>Format for</label></td>
<td>
<div class="fi">
<input name="RUN_PSIBLAST_FORM" id="RUN_PSIBLAST" type="checkbox" class="cb psiBlast hidden" />
<label for="I_THRESH">PSI-BLAST with inclusion threshold:</label>
<input name="I_THRESH" id="I_THRESH" size="10" type="text" value="" defVal="0.005" />
<a class="helplink ui-ncbitoggler" data-jig="ncbitoggler" title="PSI BLAST formatting help" id="psiHelp" href="#"><i class="fas fa-question-circle"></i><span class="usa-sr-only">Help</span></a>
<div class="ui-helper-reset" aria-live="assertive" >
<ul class="helpbox ui-ncbitoggler-slave" id="hlp9">
<li>Format for PSI-BLAST: The Position-Specific Iterated BLAST (PSI-BLAST) program performs iterative searches with a protein query,
in which sequences found in one round of search are used to build a custom score model for the next round.
<a href="https://blast.ncbi.nlm.nih.gov/doc/blast-topics/resultformatoptions.html#format-for-psi-blast" target="helpWin" title="Additional PSI BLAST formatting help">more...</a>
</li>
<li>Inclusion Threshold: This sets the statistical significance threshold for including a sequence in the model used
by PSI-BLAST to create the PSSM on the next iteration.</li>
</ul>
</div><!-- ARIA -->
</div>
</td>
</tr>
</table>
</dd>
</dl>
<input name="RID" value="KDJMCZ64013" type="hidden" />
<input name="CDD_RID" value="" type="hidden" />
<input name="CDD_SEARCH_STATE" type="hidden" value="" />
<input name="STEP_NUMBER" value="" id="stepNumber" type="hidden" />
<input name="CMD" value="Get" type="hidden" />
<input name="FORMAT_EQ_OP" value="AND" type="hidden" />
<input name="RESULTS_PAGE_TARGET" type="hidden" id="resPageTarget" value="Blast_Results_for_1792321444" />
<input name="QUERY_INFO" type="hidden" value="Protein Sequence" />
<input name="ENTREZ_QUERY" type="hidden" value="" />
<input name="QUERY_INDEX" type="hidden" value="0"/>
<input name="NUM_QUERIES" type="hidden" value="1"/>
<input name="CONFIG_DESCR" type="hidden" value="ClustMemNbr,ClustComn,Ds,Sc,Ms,Ts,Cov,Eval,Idnt,AccLen,Acc" />
<!-- Those params are set in the template (blastn.dat, blastp.dat etc. -->
<input name="BLAST_PROGRAMS" type="hidden" value="blastp"/>
<input name="PAGE" type="hidden" value="Proteins"/>
<input name="PROGRAM" type="hidden" value="blastp"/>
<input name="MEGABLAST" type="hidden" value="" />
<input name="RUN_PSIBLAST" type="hidden" value="" />
<input name="BLAST_SPEC" id="blastSpec" type="hidden" value=""/>
<input name="QUERY" type="hidden" value=""/>
<input name="JOB_TITLE" type="hidden" value="Protein Sequence"/>
<input name="QUERY_TO" type="hidden" value=""/>
<input name="QUERY_FROM" type="hidden" value=""/>
<input name="SUBJECTS_FROM" type="hidden" value=""/>
<input name="SUBJECTS_TO" type="hidden" value=""/>
<input name="EQ_TEXT" type="hidden" value=""/>
<input name="ORGN" type="hidden" value=""/>
<input name="EQ_MENU" type="hidden" value=""/>
<input name="ORG_EXCLUDE" type="hidden" value=""/>
<input name="PHI_PATTERN" type="hidden" value=""/>
<input name="EXPECT" type="hidden" value=""/>
<input name="DATABASE" type="hidden" value="nr"/>
<input name="DB_GROUP" type="hidden" value=""/>
<input name="SUBGROUP_NAME" type="hidden" value=""/>
<input name="GENETIC_CODE" type="hidden" value=""/>
<input name="WORD_SIZE" type="hidden" value="5"/>
<input name="MATCH_SCORES" type="hidden" value=""/>
<input name="MATRIX_NAME" type="hidden" value=""/>
<input name="GAPCOSTS" type="hidden" value=""/>
<input name="MAX_NUM_SEQ" id="maxNumSeq" type="hidden" value=""/>
<input name="COMPOSITION_BASED_STATISTICS" type="hidden" value=""/>
<input name="NEWWIN" type="hidden" value=""/>
<input name="SHORT_QUERY_ADJUST" type="hidden" value=""/>
<input name="FILTER" type="hidden" value=""/>
<input name="REPEATS" type="hidden" value=""/>
<input name="ID_FOR_PSSM" type="hidden" value=""/>
<input name="EXCLUDE_MODELS" type="hidden" value=""/>
<input name="EXCLUDE_SEQ_UNCULT" type="hidden" value=""/>
<input name="WP_PROTEINS" type="hidden" value=""/>
<input name="SEQ_FROM_TYPE" type="hidden" value=""/>
<input name="ENTREZ_QUERY" type="hidden" value=""/>
<input name="ENTREZ_QUERY_PRESET" type="hidden" value=""/>
<input name="ENTREZ_QUERY_PRESET_EXCL" type="hidden" value=""/>
<input name="NUM_ORG" type="hidden" value = "1" />
<!-- PSSM -->
<input name="LCASE_MASK" type="hidden" value=""/>
<input name="TEMPLATE_TYPE" type="hidden" value=""/>
<input name="TEMPLATE_LENGTH" type="hidden" value=""/>
<input name="I_THRESH" type="hidden" value=""/>
<input name="PSI_PSEUDOCOUNT" type="hidden" value=""/>
<input name="DI_THRESH" type="hidden" id="diThresh" value=""/>
<input name="HSP_RANGE_MAX" type="hidden" value=""/>
<input name="ADJUSTED_FOR_SHORT_QUERY" type="hidden" value=""/>
<input name="MIXED_QUERIES" type="hidden" value=""/>
<input name="MIXED_DATABASE" id="mixedDb" type="hidden" value=""/>
<input name="BUILD_NAME" type="hidden" value=""/>
<input name="ORG_DBS" type="hidden" value="giless_dbvers5"/>
<input name="WWW_BLAST_TYPE" type="hidden" value=""/>
<!--QBlastInfoBegin
RID = KDJMCZ64013
RTOE = 21
QBlastInfoEnd
-->
</form>
</div><!-- /#content -->
</div><!-- /#content-wrap -->
<footer>
<section class="icon-section">
<div id="icon-section-header" class="icon-section_header">Follow NCBI</div>
<div class="grid-container container">
<div class="icon-section_container">
<a class="footer-icon" id="footer_twitter" href="https://twitter.com/ncbi" aria-label="Twitter"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<defs>
<style>
.cls-11 {
fill: #737373;
}
</style>
</defs>
<title>Twitter</title>
<path class="cls-11" d="M250.11,105.48c-7,3.14-13,3.25-19.27.14,8.12-4.86,8.49-8.27,11.43-17.46a78.8,78.8,0,0,1-25,9.55,39.35,39.35,0,0,0-67,35.85,111.6,111.6,0,0,1-81-41.08A39.37,39.37,0,0,0,81.47,145a39.08,39.08,0,0,1-17.8-4.92c0,.17,0,.33,0,.5a39.32,39.32,0,0,0,31.53,38.54,39.26,39.26,0,0,1-17.75.68,39.37,39.37,0,0,0,36.72,27.3A79.07,79.07,0,0,1,56,223.34,111.31,111.31,0,0,0,116.22,241c72.3,0,111.83-59.9,111.83-111.84,0-1.71,0-3.4-.1-5.09C235.62,118.54,244.84,113.37,250.11,105.48Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_facebook" href="https://www.facebook.com/ncbi.nlm" aria-label="Facebook"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<title>Facebook</title>
<path class="cls-11" d="M210.5,115.12H171.74V97.82c0-8.14,5.39-10,9.19-10h27.14V52l-39.32-.12c-35.66,0-42.42,26.68-42.42,43.77v19.48H99.09v36.32h27.24v109h45.41v-109h35Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_linkedin" href="https://www.linkedin.com/company/ncbinlm" aria-label="LinkedIn"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<title>LinkedIn</title>
<path class="cls-11" d="M101.64,243.37H57.79v-114h43.85Zm-22-131.54h-.26c-13.25,0-21.82-10.36-21.82-21.76,0-11.65,8.84-21.15,22.33-21.15S101.7,78.72,102,90.38C102,101.77,93.4,111.83,79.63,111.83Zm100.93,52.61A17.54,17.54,0,0,0,163,182v61.39H119.18s.51-105.23,0-114H163v13a54.33,54.33,0,0,1,34.54-12.66c26,0,44.39,18.8,44.39,55.29v58.35H198.1V182A17.54,17.54,0,0,0,180.56,164.44Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_github" href="https://github.com/ncbi" aria-label="GitHub"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<defs>
<style>
.cls-11,
.cls-12 {
fill: #737373;
}
.cls-11 {
fill-rule: evenodd;
}
</style>
</defs>
<title>GitHub</title>
<path class="cls-11" d="M151.36,47.28a105.76,105.76,0,0,0-33.43,206.1c5.28,1,7.22-2.3,7.22-5.09,0-2.52-.09-10.85-.14-19.69-29.42,6.4-35.63-12.48-35.63-12.48-4.81-12.22-11.74-15.47-11.74-15.47-9.59-6.56.73-6.43.73-6.43,10.61.75,16.21,10.9,16.21,10.9,9.43,16.17,24.73,11.49,30.77,8.79,1-6.83,3.69-11.5,6.71-14.14C108.57,197.1,83.88,188,83.88,147.51a40.92,40.92,0,0,1,10.9-28.39c-1.1-2.66-4.72-13.42,1-28,0,0,8.88-2.84,29.09,10.84a100.26,100.26,0,0,1,53,0C198,88.3,206.9,91.14,206.9,91.14c5.76,14.56,2.14,25.32,1,28a40.87,40.87,0,0,1,10.89,28.39c0,40.62-24.74,49.56-48.29,52.18,3.79,3.28,7.17,9.71,7.17,19.58,0,14.15-.12,25.54-.12,29,0,2.82,1.9,6.11,7.26,5.07A105.76,105.76,0,0,0,151.36,47.28Z">
</path>
<path class="cls-12" d="M85.66,199.12c-.23.52-1.06.68-1.81.32s-1.2-1.06-.95-1.59,1.06-.69,1.82-.33,1.21,1.07.94,1.6Zm-1.3-1">
</path>
<path class="cls-12" d="M90,203.89c-.51.47-1.49.25-2.16-.49a1.61,1.61,0,0,1-.31-2.19c.52-.47,1.47-.25,2.17.49s.82,1.72.3,2.19Zm-1-1.08">
</path>
<path class="cls-12" d="M94.12,210c-.65.46-1.71,0-2.37-.91s-.64-2.07,0-2.52,1.7,0,2.36.89.65,2.08,0,2.54Zm0,0"></path>
<path class="cls-12" d="M99.83,215.87c-.58.64-1.82.47-2.72-.41s-1.18-2.06-.6-2.7,1.83-.46,2.74.41,1.2,2.07.58,2.7Zm0,0">
</path>
<path class="cls-12" d="M107.71,219.29c-.26.82-1.45,1.2-2.64.85s-2-1.34-1.74-2.17,1.44-1.23,2.65-.85,2,1.32,1.73,2.17Zm0,0">
</path>
<path class="cls-12" d="M116.36,219.92c0,.87-1,1.59-2.24,1.61s-2.29-.68-2.3-1.54,1-1.59,2.26-1.61,2.28.67,2.28,1.54Zm0,0">
</path>
<path class="cls-12" d="M124.42,218.55c.15.85-.73,1.72-2,1.95s-2.37-.3-2.52-1.14.73-1.75,2-2,2.37.29,2.53,1.16Zm0,0"></path>
</svg></a>
<a class="footer-icon" id="footer_blog" href="https://ncbiinsights.ncbi.nlm.nih.gov/" aria-label="Blog">
<svg id="Layer_1" data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 40 40"><defs><style>.cls-1{fill:#737373;}</style></defs><path class="cls-1" d="M14,30a4,4,0,1,1-4-4,4,4,0,0,1,4,4Zm11,3A19,19,0,0,0,7.05,15a1,1,0,0,0-1,1v3a1,1,0,0,0,.93,1A14,14,0,0,1,20,33.07,1,1,0,0,0,21,34h3a1,1,0,0,0,1-1Zm9,0A28,28,0,0,0,7,6,1,1,0,0,0,6,7v3a1,1,0,0,0,1,1A23,23,0,0,1,29,33a1,1,0,0,0,1,1h3A1,1,0,0,0,34,33Z"></path></svg>
</a>
</div>
</div>
</section>
<section class="container-fluid bg-primary">
<div class="container pt-5">
<div class="row mt-3">
<div class="col-lg-3 col-12">
<p><a class="text-white" href="https://www.nlm.nih.gov/socialmedia/index.html">Connect with NLM</a></p>
<ul class="list-inline social_media">
<li class="list-inline-item"><a href="https://twitter.com/NLM_NIH" aria-label="Twitter" target="_blank" rel="noopener noreferrer"><svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<style type="text/css">
.st20 {
fill: #FFFFFF;
}
.st30 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
</style>
<title>SM-Twitter</title>
<g>
<g>
<g>
<path class="st20" d="M192.9,88.1c-5,2.2-9.2,2.3-13.6,0.1c5.7-3.4,6-5.8,8.1-12.3c-5.4,3.2-11.4,5.5-17.6,6.7
c-10.5-11.2-28.1-11.7-39.2-1.2c-7.2,6.8-10.2,16.9-8,26.5c-22.3-1.1-43.1-11.7-57.2-29C58,91.6,61.8,107.9,74,116
c-4.4-0.1-8.7-1.3-12.6-3.4c0,0.1,0,0.2,0,0.4c0,13.2,9.3,24.6,22.3,27.2c-4.1,1.1-8.4,1.3-12.5,0.5c3.6,11.3,14,19,25.9,19.3
c-11.6,9.1-26.4,13.2-41.1,11.5c12.7,8.1,27.4,12.5,42.5,12.5c51,0,78.9-42.2,78.9-78.9c0-1.2,0-2.4-0.1-3.6
C182.7,97.4,189.2,93.7,192.9,88.1z"></path>
</g>
</g>
<circle class="st30" cx="124.4" cy="128.8" r="108.2"></circle>
</g>
</svg></a></li>
<li class="list-inline-item"><a href="https://www.facebook.com/nationallibraryofmedicine" aria-label="Facebook" rel="noopener noreferrer" target="_blank">
<svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<style type="text/css">
.st10 {
fill: #FFFFFF;
}
.st110 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
</style>
<title>SM-Facebook</title>
<g>
<g>
<path class="st10" d="M159,99.1h-24V88.4c0-5,3.3-6.2,5.7-6.2h16.8V60l-24.4-0.1c-22.1,0-26.2,16.5-26.2,27.1v12.1H90v22.5h16.9
v67.5H135v-67.5h21.7L159,99.1z"></path>
</g>
</g>
<circle class="st110" cx="123.6" cy="123.2" r="108.2"></circle>
</svg>
</a></li>
<li class="list-inline-item"><a href="https://www.youtube.com/user/NLMNIH" aria-label="Youtube" target="_blank" rel="noopener noreferrer"><svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<title>SM-Youtube</title>
<style type="text/css">
.st4 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
.st5 {
fill: #FFFFFF;
}
</style>
<circle class="st4" cx="124.2" cy="123.4" r="108.2"></circle>
<g transform="translate(0,-952.36218)">
<path class="st5" d="M88.4,1037.4c-10.4,0-18.7,8.3-18.7,18.7v40.1c0,10.4,8.3,18.7,18.7,18.7h72.1c10.4,0,18.7-8.3,18.7-18.7
v-40.1c0-10.4-8.3-18.7-18.7-18.7H88.4z M115.2,1058.8l29.4,17.4l-29.4,17.4V1058.8z"></path>
</g>
</svg></a></li>
</ul>
</div>
<div class="col-lg-3 col-12">
<p class="address_footer text-white">National Library of Medicine<br>
<a href="https://www.google.com/maps/place/8600+Rockville+Pike,+Bethesda,+MD+20894/@38.9959508,-77.101021,17z/data=!3m1!4b1!4m5!3m4!1s0x89b7c95e25765ddb:0x19156f88b27635b8!8m2!3d38.9959508!4d-77.0988323" class="text-white" target="_blank" rel="noopener noreferrer">8600 Rockville Pike<br>
Bethesda, MD 20894</a></p>
</div>
<div class="col-lg-3 col-12 centered-lg">
<p><a href="https://www.nlm.nih.gov/web_policies.html" class="text-white">Web Policies</a><br>
<a href="https://www.nih.gov/institutes-nih/nih-office-director/office-communications-public-liaison/freedom-information-act-office" class="text-white">FOIA</a><br>
<a href="https://www.hhs.gov/vulnerability-disclosure-policy/index.html" class="text-white" id="vdp">HHS Vulnerability Disclosure</a></p>
</div>
<div class="col-lg-3 col-12 centered-lg">
<p><a class="supportLink text-white" href="https://support.nlm.nih.gov/">Help</a><br>
<a href="https://www.nlm.nih.gov/accessibility.html" class="text-white">Accessibility</a><br>
<a href="https://www.nlm.nih.gov/careers/careers.html" class="text-white">Careers</a></p>
</div>
</div>
<div class="row">
<div class="col-lg-12 centered-lg">
<nav class="bottom-links">
<ul class="mt-3">
<li>
<a class="text-white" href="//www.nlm.nih.gov/">NLM</a>
</li>
<li>
<a class="text-white" href="https://www.nih.gov/">NIH</a>
</li>
<li>
<a class="text-white" href="https://www.hhs.gov/">HHS</a>
</li>
<li>
<a class="text-white" href="https://www.usa.gov/">USA.gov</a>
</li>
</ul>
</nav>
</div>
</div>
</div>
</section>
</footer>
<script type="text/javascript" src="js/nwds.js"></script>
<script type="text/javascript" src="js/ncbipopup.js"></script>
<script type="text/javascript" src="js/remote_data_provider.js?v=2"></script>
<script type="text/javascript" src="js/headerNew.js?v=2"></script>
<script src="js/uswds.min.js"></script>
</div><!--/#wrap-->
<script type="text/javascript" src="/portal/portal3rc.fcgi/rlib/js/InstrumentOmnitureBaseJS/InstrumentNCBIBaseJS/InstrumentPageStarterJS.js"></script>
<!--
<script type="text/javascript" src="://www.ncbi.nlm.nih.gov/portal/portal3rc.fcgi/supportedbrowsers/js/nonportalbc_min.js"></script>
<script type="text/javascript">$("#browsers_ajax").browserCheck();</script>
-->
</body>
</html>
BLAST GET¶
Using the request id parsed from the result of the PUT, get the results of the search. Common attributes:
- RID - mandatory
- FORMAT_TYPE - HTML, Text, ASN.1, XML
- ALIGNMENTS - number of alignments (default 500)
- ALIGNMENT_VIEW - Pairwise, QueryAnchored, QueryAnchoredNoIdentities, FlatQueryAnchored, FlatQueryAnchoredNoIdentities, Tabular
rid = re.search(r'RID = (\S+)',response.text).group(1)
print(rid)
KDJMCZ64013
result = requests.get('http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=GET&RID=%s&FORMAT_TYPE=XML' % rid).text
print(result)
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" lang="en">
<head>
<meta http-equiv="Content-Type" content="text/html; charset=UTF-8" />
<meta name="jig" content="ncbitoggler"/>
<meta name="ncbitoggler" content="animation:'none'"/>
<title>NCBI Blast:</title>
<script type="text/javascript" src="/core/jig/1.15.2/js/jig.min.js "></script>
<script type="text/javascript"> jQuery.getScript("/core/alerts/alerts.js", function() {
galert(['div#header', 'body > *:nth-child(1)'])
});</script>
<meta http-equiv="Pragma" content="no-cache">
<link rel="stylesheet" type="text/css" href="css/uswds.min.css" media="screen" />
<link rel="stylesheet" type="text/css" href="https://www.ncbi.nlm.nih.gov/style-guide/static/nwds/css/nwds.css"/>
<link rel="stylesheet" href="css/headerNew.css?v=1"/>
<link rel="stylesheet" href="https://use.fontawesome.com/releases/v5.5.0/css/all.css" crossorigin="anonymous"> <!-- Font Awesome icons -->
<link rel="stylesheet" type="text/css" href="css/footerNew.css?v=1" media="screen" />
<link rel="stylesheet" type="text/css" href="css/main.css" media="screen" />
<link rel="stylesheet" type="text/css" href="css/blastRes.css" media="screen" />
<link rel="stylesheet" type="text/css" href="css/print.css" media="print" />
<!--[if lte IE 6]>
<link rel="stylesheet" type="text/css" href="css/ie6_or_less.css" />
<![endif]-->
<script type="text/javascript" src="js/utils.js"></script>
<script type="text/javascript" src="js/results.js"></script>
</head>
<body id="type-a" class="noToggleCheck" >
<div id="wrap">
<script>var useOfficialGovtHeader = true;</script>
<section class="usa-banner">
<div class="usa-accordion">
<header class="usa-banner-header">
<div class="usa-grid usa-banner-inner">
<img src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/favicons/favicon-57.png" alt="U.S. flag">
<p>An official website of the United States government</p>
<button class="usa-accordion-button usa-banner-button" aria-expanded="false" aria-controls="gov-banner-top">
<span class="usa-banner-button-text">Here's how you know</span>
</button>
</div>
</header>
<div class="usa-banner-content usa-grid usa-accordion-content" id="gov-banner-top" aria-hidden="true">
<div class="usa-banner-guidance-gov usa-width-one-half">
<img class="usa-banner-icon usa-media_block-img" src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/icon-dot-gov.svg" alt="Dot gov">
<div class="usa-media_block-body">
<p>
<strong>The .gov means itâs official.</strong>
<br>
Federal government websites often end in .gov or .mil. Before
sharing sensitive information, make sure youâre on a federal
government site.
</p>
</div>
</div>
<div class="usa-banner-guidance-ssl usa-width-one-half">
<img class="usa-banner-icon usa-media_block-img" src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/icon-https.svg" alt="Https">
<div class="usa-media_block-body">
<p>
<strong>The site is secure.</strong>
<br>
The <strong>https://</strong> ensures that you are connecting to the
official website and that any information you provide is encrypted
and transmitted securely.
</p>
</div>
</div>
</div>
</div>
</section>
<header class="ncbi-header" role="banner" data-section="Header">
<a class="usa-skipnav" href="#mainCont">Skip to main page content</a>
<div class="usa-grid">
<div class="usa-width-one-whole">
<div class="ncbi-header__logo">
<a href="https://www.ncbi.nlm.nih.gov/" class="logo" aria-label="NCBI Logo" data-ga-action="click_image" data-ga-label="NIH NLM Logo">
<img src="https://www.ncbi.nlm.nih.gov/coreutils/nwds/img/logos/AgencyLogo.svg" alt="NIH NLM Logo">
</a>
</div>
<div class="ncbi-header__account">
<a id="account_login" href="https://www.ncbi.nlm.nih.gov/account/?back_url=https%3A%2F%2Fblast%2Encbi%2Enlm%2Enih%2Egov%2FBlast%2Ecgi%3FCMD%3DGET%26FORMAT%5FTYPE%3DXML%26RID%3DKDJMCZ64013" class="usa-button header-button">Log in</a>
<button id="account_info" class="header-button" aria-controls="account_popup">
<span class="fa fa-user" aria-hidden="true"></span>
<span class="username desktop-only" aria-hidden="true" id="uname_short"></span>
<span class="sr-only">Show account info</span>
</button>
</div>
<div class="ncbi-popup-anchor">
<div class="ncbi-popup account-popup" id="account_popup" aria-hidden="true" role="dialog" aria-labelledby="account-popup-header">
<div class="ncbi-popup-head">
<button class="ncbi-close-button"><span class="fa fa-window-close"></span><span class="usa-sr-only">Close</span></button>
<h4>Account</h4>
</div>
<div class="account-user-info">
Logged in as:<br>
<b><span class="username" id="uname_long">username</span></b>
</div>
<div class="account-links">
<ul class="usa-unstyled-list">
<li><a id="account_myncbi" href="https://www.ncbi.nlm.nih.gov/myncbi/">Dashboard</a> <span class="ncbi-text-small-light">(My NCBI)</span></li>
<li><a id="account_pubs" href="https://www.ncbi.nlm.nih.gov/myncbi/collections/bibliography/">Publications</a> <span class="ncbi-text-small-light">(My Bibliography)</span></li>
<li><a id="account_settings" href="https://www.ncbi.nlm.nih.gov/account/settings/">Account settings</a></li>
<li><a id="account_logout" href="https://www.ncbi.nlm.nih.gov/account/signout/?back_url=https%3A%2F%2Fblast%2Encbi%2Enlm%2Enih%2Egov%2FBlast%2Ecgi%3FCMD%3DGET%26FORMAT%5FTYPE%3DXML%26RID%3DKDJMCZ64013">Log out</a></li>
</ul>
</div>
</div>
</div>
</div>
</div>
</header>
<div role="navigation" aria-label="access keys">
<a id="nws_header_accesskey_0" href="https://www.ncbi.nlm.nih.gov/guide/browsers/#ncbi_accesskeys" class="usa-sr-only" accesskey="0" tabindex="-1">Access keys</a>
<a id="nws_header_accesskey_1" href="https://www.ncbi.nlm.nih.gov" class="usa-sr-only" accesskey="1" tabindex="-1">NCBI Homepage</a>
<a id="nws_header_accesskey_2" href="/myncbi/" class="set-base-url usa-sr-only" accesskey="2" tabindex="-1">MyNCBI Homepage</a>
<a id="nws_header_accesskey_3" href="#maincontent" class="usa-sr-only" accesskey="3" tabindex="-1">Main Content</a>
<a id="nws_header_accesskey_4" href="#" class="usa-sr-only" accesskey="4" tabindex="-1">Main Navigation</a>
</div>
<nav class="ncbi-topnav" id="navcontent">
<div class="usa-grid">
<a class="ncbi-topnav-root" href="Blast.cgi">BLAST <sup>®</sup></a> <span id="brc"><span class="brcrmbsign">»</span> <a href="Blast.cgi?PAGE=Nucleotides&PROGRAM=blastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=">blastn suite</a> <span class="brcrmbsign">»</span> RID-KDJMCZ64013</span>
<ul class="rf ncbi-topnav-list" id="topnav-list">
<li class="first "><a href="Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome" title="BLAST Home">Home</a></li>
<li class="recent "><a href="Blast.cgi?CMD=GetSaved&RECENT_RESULTS=on" title="Unexpired BLAST jobs">Recent Results</a></li>
<li class="saved "><a href="Blast.cgi?CMD=GetSaved" title="Saved sets of BLAST search parameters">Saved Strategies</a></li>
<li class= "last documentation "> <a href="../doc/blast-help/" title="BLAST documentation">Help</a></li>
</ul>
</div>
</nav>
<div id="content-wrap">
<div class="pageTitle">
Format Request Status
</div>
<div class="inlineDiv resHeader">
<a id="frmPage" class="WAITING" href="#" submitForm="reformat">[Formatting options] </a>
</div>
<h2 id="jtitle" >Job Title: </h2>
<div id="content">
<!--<ul id="msg" class="msg"><li class=""><p class=""></p><p class=""></p><p class=""></p></ul> -->
<ul id="msg" class="msg"><li class=""></li></ul>
<p><!--
QBlastInfoBegin
Status=WAITING
QBlastInfoEnd
--></p>
<SCRIPT LANGUAGE="JavaScript"><!--
var tm = "2000";
if (tm != "") {
setTimeout('document.forms[0].submit();',tm);
}
//--></SCRIPT>
<table id="statInfo" class="WAITING">
<tr><td>Request ID</td><td> <b>KDJMCZ64013</b></td></tr>
<tr class="odd"><td>Status</td><td>Searching</td></tr>
<tr><td>Submitted at</td><td>Mon Oct 23 15:33:32 2023</td></tr>
<tr class="odd"><td>Current time</td><td>Mon Oct 23 15:33:52 2023</td></tr>
<tr><td>Time since submission</td><td>00:00:20</td></tr>
</table>
<p class="WAITING">This page will be automatically updated in <b>2</b> seconds</p>
<form action="Blast.cgi" enctype="application/x-www-form-urlencoded" method="POST" id="results">
<input name="FORMAT_TYPE" type="hidden" value="XML"><input name="RID" type="hidden" value="KDJMCZ64013"><input name="SEARCH_DB_STATUS" type="hidden" value="31"><input name="USER_TYPE" type="hidden" value="2"><input name="_PGR" type="hidden" value="0">
<input name="_PGR" type="hidden" value="0" >
<input name="CMD" type="hidden" value="Get">
</form>
</div><!-- /#content -->
<form action="Blast.cgi" enctype="application/x-www-form-urlencoded" method="post" name="reformat" id="reformat">
<input name="QUERY_INFO" type="hidden" value="" />
<input name="ENTREZ_QUERY" type="hidden" value="" />
<input name="CDD_RID" type="hidden" value="" />
<input name="CDD_SEARCH_STATE" type="hidden" value="" />
<input name="RID" type="hidden" value="KDJMCZ64013" />
<input name="STEP_NUMBER" type="hidden" value="" />
<input name="CMD" type="hidden" value="Web"/>
<input NAME="PAGE_TYPE" type="hidden" value="BlastFormatting"/>
<!-- TO DO: test all of those changes -->
<!-- Psi blast params PSI_BLAST_PARAMS - commented- using forms[0] from fromatter> -->
<!-- Current Formatting options FORMATTING_OPTIONS- commented- using forms[0] from fromatter> -->
<!-- Current Search options CURR_SAVED_OPTIONS - commented- using forms[0] from fromatter> -->
</form>
</div><!-- /#content-wrap -->
<footer>
<section class="icon-section">
<div id="icon-section-header" class="icon-section_header">Follow NCBI</div>
<div class="grid-container container">
<div class="icon-section_container">
<a class="footer-icon" id="footer_twitter" href="https://twitter.com/ncbi" aria-label="Twitter"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<defs>
<style>
.cls-11 {
fill: #737373;
}
</style>
</defs>
<title>Twitter</title>
<path class="cls-11" d="M250.11,105.48c-7,3.14-13,3.25-19.27.14,8.12-4.86,8.49-8.27,11.43-17.46a78.8,78.8,0,0,1-25,9.55,39.35,39.35,0,0,0-67,35.85,111.6,111.6,0,0,1-81-41.08A39.37,39.37,0,0,0,81.47,145a39.08,39.08,0,0,1-17.8-4.92c0,.17,0,.33,0,.5a39.32,39.32,0,0,0,31.53,38.54,39.26,39.26,0,0,1-17.75.68,39.37,39.37,0,0,0,36.72,27.3A79.07,79.07,0,0,1,56,223.34,111.31,111.31,0,0,0,116.22,241c72.3,0,111.83-59.9,111.83-111.84,0-1.71,0-3.4-.1-5.09C235.62,118.54,244.84,113.37,250.11,105.48Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_facebook" href="https://www.facebook.com/ncbi.nlm" aria-label="Facebook"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<title>Facebook</title>
<path class="cls-11" d="M210.5,115.12H171.74V97.82c0-8.14,5.39-10,9.19-10h27.14V52l-39.32-.12c-35.66,0-42.42,26.68-42.42,43.77v19.48H99.09v36.32h27.24v109h45.41v-109h35Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_linkedin" href="https://www.linkedin.com/company/ncbinlm" aria-label="LinkedIn"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<title>LinkedIn</title>
<path class="cls-11" d="M101.64,243.37H57.79v-114h43.85Zm-22-131.54h-.26c-13.25,0-21.82-10.36-21.82-21.76,0-11.65,8.84-21.15,22.33-21.15S101.7,78.72,102,90.38C102,101.77,93.4,111.83,79.63,111.83Zm100.93,52.61A17.54,17.54,0,0,0,163,182v61.39H119.18s.51-105.23,0-114H163v13a54.33,54.33,0,0,1,34.54-12.66c26,0,44.39,18.8,44.39,55.29v58.35H198.1V182A17.54,17.54,0,0,0,180.56,164.44Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_github" href="https://github.com/ncbi" aria-label="GitHub"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<defs>
<style>
.cls-11,
.cls-12 {
fill: #737373;
}
.cls-11 {
fill-rule: evenodd;
}
</style>
</defs>
<title>GitHub</title>
<path class="cls-11" d="M151.36,47.28a105.76,105.76,0,0,0-33.43,206.1c5.28,1,7.22-2.3,7.22-5.09,0-2.52-.09-10.85-.14-19.69-29.42,6.4-35.63-12.48-35.63-12.48-4.81-12.22-11.74-15.47-11.74-15.47-9.59-6.56.73-6.43.73-6.43,10.61.75,16.21,10.9,16.21,10.9,9.43,16.17,24.73,11.49,30.77,8.79,1-6.83,3.69-11.5,6.71-14.14C108.57,197.1,83.88,188,83.88,147.51a40.92,40.92,0,0,1,10.9-28.39c-1.1-2.66-4.72-13.42,1-28,0,0,8.88-2.84,29.09,10.84a100.26,100.26,0,0,1,53,0C198,88.3,206.9,91.14,206.9,91.14c5.76,14.56,2.14,25.32,1,28a40.87,40.87,0,0,1,10.89,28.39c0,40.62-24.74,49.56-48.29,52.18,3.79,3.28,7.17,9.71,7.17,19.58,0,14.15-.12,25.54-.12,29,0,2.82,1.9,6.11,7.26,5.07A105.76,105.76,0,0,0,151.36,47.28Z">
</path>
<path class="cls-12" d="M85.66,199.12c-.23.52-1.06.68-1.81.32s-1.2-1.06-.95-1.59,1.06-.69,1.82-.33,1.21,1.07.94,1.6Zm-1.3-1">
</path>
<path class="cls-12" d="M90,203.89c-.51.47-1.49.25-2.16-.49a1.61,1.61,0,0,1-.31-2.19c.52-.47,1.47-.25,2.17.49s.82,1.72.3,2.19Zm-1-1.08">
</path>
<path class="cls-12" d="M94.12,210c-.65.46-1.71,0-2.37-.91s-.64-2.07,0-2.52,1.7,0,2.36.89.65,2.08,0,2.54Zm0,0"></path>
<path class="cls-12" d="M99.83,215.87c-.58.64-1.82.47-2.72-.41s-1.18-2.06-.6-2.7,1.83-.46,2.74.41,1.2,2.07.58,2.7Zm0,0">
</path>
<path class="cls-12" d="M107.71,219.29c-.26.82-1.45,1.2-2.64.85s-2-1.34-1.74-2.17,1.44-1.23,2.65-.85,2,1.32,1.73,2.17Zm0,0">
</path>
<path class="cls-12" d="M116.36,219.92c0,.87-1,1.59-2.24,1.61s-2.29-.68-2.3-1.54,1-1.59,2.26-1.61,2.28.67,2.28,1.54Zm0,0">
</path>
<path class="cls-12" d="M124.42,218.55c.15.85-.73,1.72-2,1.95s-2.37-.3-2.52-1.14.73-1.75,2-2,2.37.29,2.53,1.16Zm0,0"></path>
</svg></a>
<a class="footer-icon" id="footer_blog" href="https://ncbiinsights.ncbi.nlm.nih.gov/" aria-label="Blog">
<svg id="Layer_1" data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 40 40"><defs><style>.cls-1{fill:#737373;}</style></defs><path class="cls-1" d="M14,30a4,4,0,1,1-4-4,4,4,0,0,1,4,4Zm11,3A19,19,0,0,0,7.05,15a1,1,0,0,0-1,1v3a1,1,0,0,0,.93,1A14,14,0,0,1,20,33.07,1,1,0,0,0,21,34h3a1,1,0,0,0,1-1Zm9,0A28,28,0,0,0,7,6,1,1,0,0,0,6,7v3a1,1,0,0,0,1,1A23,23,0,0,1,29,33a1,1,0,0,0,1,1h3A1,1,0,0,0,34,33Z"></path></svg>
</a>
</div>
</div>
</section>
<section class="container-fluid bg-primary">
<div class="container pt-5">
<div class="row mt-3">
<div class="col-lg-3 col-12">
<p><a class="text-white" href="https://www.nlm.nih.gov/socialmedia/index.html">Connect with NLM</a></p>
<ul class="list-inline social_media">
<li class="list-inline-item"><a href="https://twitter.com/NLM_NIH" aria-label="Twitter" target="_blank" rel="noopener noreferrer"><svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<style type="text/css">
.st20 {
fill: #FFFFFF;
}
.st30 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
</style>
<title>SM-Twitter</title>
<g>
<g>
<g>
<path class="st20" d="M192.9,88.1c-5,2.2-9.2,2.3-13.6,0.1c5.7-3.4,6-5.8,8.1-12.3c-5.4,3.2-11.4,5.5-17.6,6.7
c-10.5-11.2-28.1-11.7-39.2-1.2c-7.2,6.8-10.2,16.9-8,26.5c-22.3-1.1-43.1-11.7-57.2-29C58,91.6,61.8,107.9,74,116
c-4.4-0.1-8.7-1.3-12.6-3.4c0,0.1,0,0.2,0,0.4c0,13.2,9.3,24.6,22.3,27.2c-4.1,1.1-8.4,1.3-12.5,0.5c3.6,11.3,14,19,25.9,19.3
c-11.6,9.1-26.4,13.2-41.1,11.5c12.7,8.1,27.4,12.5,42.5,12.5c51,0,78.9-42.2,78.9-78.9c0-1.2,0-2.4-0.1-3.6
C182.7,97.4,189.2,93.7,192.9,88.1z"></path>
</g>
</g>
<circle class="st30" cx="124.4" cy="128.8" r="108.2"></circle>
</g>
</svg></a></li>
<li class="list-inline-item"><a href="https://www.facebook.com/nationallibraryofmedicine" aria-label="Facebook" rel="noopener noreferrer" target="_blank">
<svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<style type="text/css">
.st10 {
fill: #FFFFFF;
}
.st110 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
</style>
<title>SM-Facebook</title>
<g>
<g>
<path class="st10" d="M159,99.1h-24V88.4c0-5,3.3-6.2,5.7-6.2h16.8V60l-24.4-0.1c-22.1,0-26.2,16.5-26.2,27.1v12.1H90v22.5h16.9
v67.5H135v-67.5h21.7L159,99.1z"></path>
</g>
</g>
<circle class="st110" cx="123.6" cy="123.2" r="108.2"></circle>
</svg>
</a></li>
<li class="list-inline-item"><a href="https://www.youtube.com/user/NLMNIH" aria-label="Youtube" target="_blank" rel="noopener noreferrer"><svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<title>SM-Youtube</title>
<style type="text/css">
.st4 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
.st5 {
fill: #FFFFFF;
}
</style>
<circle class="st4" cx="124.2" cy="123.4" r="108.2"></circle>
<g transform="translate(0,-952.36218)">
<path class="st5" d="M88.4,1037.4c-10.4,0-18.7,8.3-18.7,18.7v40.1c0,10.4,8.3,18.7,18.7,18.7h72.1c10.4,0,18.7-8.3,18.7-18.7
v-40.1c0-10.4-8.3-18.7-18.7-18.7H88.4z M115.2,1058.8l29.4,17.4l-29.4,17.4V1058.8z"></path>
</g>
</svg></a></li>
</ul>
</div>
<div class="col-lg-3 col-12">
<p class="address_footer text-white">National Library of Medicine<br>
<a href="https://www.google.com/maps/place/8600+Rockville+Pike,+Bethesda,+MD+20894/@38.9959508,-77.101021,17z/data=!3m1!4b1!4m5!3m4!1s0x89b7c95e25765ddb:0x19156f88b27635b8!8m2!3d38.9959508!4d-77.0988323" class="text-white" target="_blank" rel="noopener noreferrer">8600 Rockville Pike<br>
Bethesda, MD 20894</a></p>
</div>
<div class="col-lg-3 col-12 centered-lg">
<p><a href="https://www.nlm.nih.gov/web_policies.html" class="text-white">Web Policies</a><br>
<a href="https://www.nih.gov/institutes-nih/nih-office-director/office-communications-public-liaison/freedom-information-act-office" class="text-white">FOIA</a><br>
<a href="https://www.hhs.gov/vulnerability-disclosure-policy/index.html" class="text-white" id="vdp">HHS Vulnerability Disclosure</a></p>
</div>
<div class="col-lg-3 col-12 centered-lg">
<p><a class="supportLink text-white" href="https://support.nlm.nih.gov/">Help</a><br>
<a href="https://www.nlm.nih.gov/accessibility.html" class="text-white">Accessibility</a><br>
<a href="https://www.nlm.nih.gov/careers/careers.html" class="text-white">Careers</a></p>
</div>
</div>
<div class="row">
<div class="col-lg-12 centered-lg">
<nav class="bottom-links">
<ul class="mt-3">
<li>
<a class="text-white" href="//www.nlm.nih.gov/">NLM</a>
</li>
<li>
<a class="text-white" href="https://www.nih.gov/">NIH</a>
</li>
<li>
<a class="text-white" href="https://www.hhs.gov/">HHS</a>
</li>
<li>
<a class="text-white" href="https://www.usa.gov/">USA.gov</a>
</li>
</ul>
</nav>
</div>
</div>
</div>
</section>
</footer>
<script type="text/javascript" src="js/nwds.js"></script>
<script type="text/javascript" src="js/ncbipopup.js"></script>
<script type="text/javascript" src="js/remote_data_provider.js?v=2"></script>
<script type="text/javascript" src="js/headerNew.js?v=2"></script>
<script src="js/uswds.min.js"></script>
<script type="text/javascript" src="/portal/portal3rc.fcgi/rlib/js/InstrumentOmnitureBaseJS/InstrumentNCBIBaseJS/InstrumentPageStarterJS.js"></script>
<!--
<script type="text/javascript" src="://www.ncbi.nlm.nih.gov/portal/portal3rc.fcgi/supportedbrowsers/js/nonportalbc_min.js"></script>
<script type="text/javascript">$("#browsers_ajax").browserCheck();</script>
-->
</div><!--/#wrap-->
</body>
</html>
result = requests.get('http://www.ncbi.nlm.nih.gov/blast/Blast.cgi?CMD=GET&RID=%s&FORMAT_TYPE=XML' % rid).text
print(result)
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml" xml:lang="en" lang="en">
<head>
<meta http-equiv="Content-Type" content="text/html; charset=UTF-8" />
<meta name="jig" content="ncbitoggler"/>
<meta name="ncbitoggler" content="animation:'none'"/>
<title>NCBI Blast:</title>
<script type="text/javascript" src="/core/jig/1.15.2/js/jig.min.js "></script>
<script type="text/javascript"> jQuery.getScript("/core/alerts/alerts.js", function() {
galert(['div#header', 'body > *:nth-child(1)'])
});</script>
<meta http-equiv="Pragma" content="no-cache">
<link rel="stylesheet" type="text/css" href="css/uswds.min.css" media="screen" />
<link rel="stylesheet" type="text/css" href="https://www.ncbi.nlm.nih.gov/style-guide/static/nwds/css/nwds.css"/>
<link rel="stylesheet" href="css/headerNew.css?v=1"/>
<link rel="stylesheet" href="https://use.fontawesome.com/releases/v5.5.0/css/all.css" crossorigin="anonymous"> <!-- Font Awesome icons -->
<link rel="stylesheet" type="text/css" href="css/footerNew.css?v=1" media="screen" />
<link rel="stylesheet" type="text/css" href="css/main.css" media="screen" />
<link rel="stylesheet" type="text/css" href="css/blastRes.css" media="screen" />
<link rel="stylesheet" type="text/css" href="css/print.css" media="print" />
<!--[if lte IE 6]>
<link rel="stylesheet" type="text/css" href="css/ie6_or_less.css" />
<![endif]-->
<script type="text/javascript" src="js/utils.js"></script>
<script type="text/javascript" src="js/results.js"></script>
</head>
<body id="type-a" class="noToggleCheck" >
<div id="wrap">
<script>var useOfficialGovtHeader = true;</script>
<section class="usa-banner">
<div class="usa-accordion">
<header class="usa-banner-header">
<div class="usa-grid usa-banner-inner">
<img src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/favicons/favicon-57.png" alt="U.S. flag">
<p>An official website of the United States government</p>
<button class="usa-accordion-button usa-banner-button" aria-expanded="false" aria-controls="gov-banner-top">
<span class="usa-banner-button-text">Here's how you know</span>
</button>
</div>
</header>
<div class="usa-banner-content usa-grid usa-accordion-content" id="gov-banner-top" aria-hidden="true">
<div class="usa-banner-guidance-gov usa-width-one-half">
<img class="usa-banner-icon usa-media_block-img" src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/icon-dot-gov.svg" alt="Dot gov">
<div class="usa-media_block-body">
<p>
<strong>The .gov means itâs official.</strong>
<br>
Federal government websites often end in .gov or .mil. Before
sharing sensitive information, make sure youâre on a federal
government site.
</p>
</div>
</div>
<div class="usa-banner-guidance-ssl usa-width-one-half">
<img class="usa-banner-icon usa-media_block-img" src="https://www.ncbi.nlm.nih.gov/coreutils/uswds/img/icon-https.svg" alt="Https">
<div class="usa-media_block-body">
<p>
<strong>The site is secure.</strong>
<br>
The <strong>https://</strong> ensures that you are connecting to the
official website and that any information you provide is encrypted
and transmitted securely.
</p>
</div>
</div>
</div>
</div>
</section>
<header class="ncbi-header" role="banner" data-section="Header">
<a class="usa-skipnav" href="#mainCont">Skip to main page content</a>
<div class="usa-grid">
<div class="usa-width-one-whole">
<div class="ncbi-header__logo">
<a href="https://www.ncbi.nlm.nih.gov/" class="logo" aria-label="NCBI Logo" data-ga-action="click_image" data-ga-label="NIH NLM Logo">
<img src="https://www.ncbi.nlm.nih.gov/coreutils/nwds/img/logos/AgencyLogo.svg" alt="NIH NLM Logo">
</a>
</div>
<div class="ncbi-header__account">
<a id="account_login" href="https://www.ncbi.nlm.nih.gov/account/?back_url=https%3A%2F%2Fblast%2Encbi%2Enlm%2Enih%2Egov%2FBlast%2Ecgi%3FCMD%3DGET%26FORMAT%5FTYPE%3DXML%26RID%3DKDJMCZ64013" class="usa-button header-button">Log in</a>
<button id="account_info" class="header-button" aria-controls="account_popup">
<span class="fa fa-user" aria-hidden="true"></span>
<span class="username desktop-only" aria-hidden="true" id="uname_short"></span>
<span class="sr-only">Show account info</span>
</button>
</div>
<div class="ncbi-popup-anchor">
<div class="ncbi-popup account-popup" id="account_popup" aria-hidden="true" role="dialog" aria-labelledby="account-popup-header">
<div class="ncbi-popup-head">
<button class="ncbi-close-button"><span class="fa fa-window-close"></span><span class="usa-sr-only">Close</span></button>
<h4>Account</h4>
</div>
<div class="account-user-info">
Logged in as:<br>
<b><span class="username" id="uname_long">username</span></b>
</div>
<div class="account-links">
<ul class="usa-unstyled-list">
<li><a id="account_myncbi" href="https://www.ncbi.nlm.nih.gov/myncbi/">Dashboard</a> <span class="ncbi-text-small-light">(My NCBI)</span></li>
<li><a id="account_pubs" href="https://www.ncbi.nlm.nih.gov/myncbi/collections/bibliography/">Publications</a> <span class="ncbi-text-small-light">(My Bibliography)</span></li>
<li><a id="account_settings" href="https://www.ncbi.nlm.nih.gov/account/settings/">Account settings</a></li>
<li><a id="account_logout" href="https://www.ncbi.nlm.nih.gov/account/signout/?back_url=https%3A%2F%2Fblast%2Encbi%2Enlm%2Enih%2Egov%2FBlast%2Ecgi%3FCMD%3DGET%26FORMAT%5FTYPE%3DXML%26RID%3DKDJMCZ64013">Log out</a></li>
</ul>
</div>
</div>
</div>
</div>
</div>
</header>
<div role="navigation" aria-label="access keys">
<a id="nws_header_accesskey_0" href="https://www.ncbi.nlm.nih.gov/guide/browsers/#ncbi_accesskeys" class="usa-sr-only" accesskey="0" tabindex="-1">Access keys</a>
<a id="nws_header_accesskey_1" href="https://www.ncbi.nlm.nih.gov" class="usa-sr-only" accesskey="1" tabindex="-1">NCBI Homepage</a>
<a id="nws_header_accesskey_2" href="/myncbi/" class="set-base-url usa-sr-only" accesskey="2" tabindex="-1">MyNCBI Homepage</a>
<a id="nws_header_accesskey_3" href="#maincontent" class="usa-sr-only" accesskey="3" tabindex="-1">Main Content</a>
<a id="nws_header_accesskey_4" href="#" class="usa-sr-only" accesskey="4" tabindex="-1">Main Navigation</a>
</div>
<nav class="ncbi-topnav" id="navcontent">
<div class="usa-grid">
<a class="ncbi-topnav-root" href="Blast.cgi">BLAST <sup>®</sup></a> <span id="brc"><span class="brcrmbsign">»</span> <a href="Blast.cgi?PAGE=Nucleotides&PROGRAM=blastn&PAGE_TYPE=BlastSearch&BLAST_SPEC=">blastn suite</a> <span class="brcrmbsign">»</span> RID-KDJMCZ64013</span>
<ul class="rf ncbi-topnav-list" id="topnav-list">
<li class="first "><a href="Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome" title="BLAST Home">Home</a></li>
<li class="recent "><a href="Blast.cgi?CMD=GetSaved&RECENT_RESULTS=on" title="Unexpired BLAST jobs">Recent Results</a></li>
<li class="saved "><a href="Blast.cgi?CMD=GetSaved" title="Saved sets of BLAST search parameters">Saved Strategies</a></li>
<li class= "last documentation "> <a href="../doc/blast-help/" title="BLAST documentation">Help</a></li>
</ul>
</div>
</nav>
<div id="content-wrap">
<div class="pageTitle">
Format Request Status
</div>
<div class="inlineDiv resHeader">
<a id="frmPage" class="WAITING" href="#" submitForm="reformat">[Formatting options] </a>
</div>
<h2 id="jtitle" >Job Title: </h2>
<div id="content">
<!--<ul id="msg" class="msg"><li class=""><p class=""></p><p class=""></p><p class=""></p></ul> -->
<ul id="msg" class="msg"><li class=""></li></ul>
<p><!--
QBlastInfoBegin
Status=WAITING
QBlastInfoEnd
--></p>
<SCRIPT LANGUAGE="JavaScript"><!--
var tm = "2000";
if (tm != "") {
setTimeout('document.forms[0].submit();',tm);
}
//--></SCRIPT>
<table id="statInfo" class="WAITING">
<tr><td>Request ID</td><td> <b>KDJMCZ64013</b></td></tr>
<tr class="odd"><td>Status</td><td>Searching</td></tr>
<tr><td>Submitted at</td><td>Mon Oct 23 15:33:32 2023</td></tr>
<tr class="odd"><td>Current time</td><td>Mon Oct 23 15:33:58 2023</td></tr>
<tr><td>Time since submission</td><td>00:00:26</td></tr>
</table>
<p class="WAITING">This page will be automatically updated in <b>2</b> seconds</p>
<form action="Blast.cgi" enctype="application/x-www-form-urlencoded" method="POST" id="results">
<input name="FORMAT_TYPE" type="hidden" value="XML"><input name="RID" type="hidden" value="KDJMCZ64013"><input name="SEARCH_DB_STATUS" type="hidden" value="31"><input name="USER_TYPE" type="hidden" value="2"><input name="_PGR" type="hidden" value="0">
<input name="_PGR" type="hidden" value="0" >
<input name="CMD" type="hidden" value="Get">
</form>
</div><!-- /#content -->
<form action="Blast.cgi" enctype="application/x-www-form-urlencoded" method="post" name="reformat" id="reformat">
<input name="QUERY_INFO" type="hidden" value="" />
<input name="ENTREZ_QUERY" type="hidden" value="" />
<input name="CDD_RID" type="hidden" value="" />
<input name="CDD_SEARCH_STATE" type="hidden" value="" />
<input name="RID" type="hidden" value="KDJMCZ64013" />
<input name="STEP_NUMBER" type="hidden" value="" />
<input name="CMD" type="hidden" value="Web"/>
<input NAME="PAGE_TYPE" type="hidden" value="BlastFormatting"/>
<!-- TO DO: test all of those changes -->
<!-- Psi blast params PSI_BLAST_PARAMS - commented- using forms[0] from fromatter> -->
<!-- Current Formatting options FORMATTING_OPTIONS- commented- using forms[0] from fromatter> -->
<!-- Current Search options CURR_SAVED_OPTIONS - commented- using forms[0] from fromatter> -->
</form>
</div><!-- /#content-wrap -->
<footer>
<section class="icon-section">
<div id="icon-section-header" class="icon-section_header">Follow NCBI</div>
<div class="grid-container container">
<div class="icon-section_container">
<a class="footer-icon" id="footer_twitter" href="https://twitter.com/ncbi" aria-label="Twitter"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<defs>
<style>
.cls-11 {
fill: #737373;
}
</style>
</defs>
<title>Twitter</title>
<path class="cls-11" d="M250.11,105.48c-7,3.14-13,3.25-19.27.14,8.12-4.86,8.49-8.27,11.43-17.46a78.8,78.8,0,0,1-25,9.55,39.35,39.35,0,0,0-67,35.85,111.6,111.6,0,0,1-81-41.08A39.37,39.37,0,0,0,81.47,145a39.08,39.08,0,0,1-17.8-4.92c0,.17,0,.33,0,.5a39.32,39.32,0,0,0,31.53,38.54,39.26,39.26,0,0,1-17.75.68,39.37,39.37,0,0,0,36.72,27.3A79.07,79.07,0,0,1,56,223.34,111.31,111.31,0,0,0,116.22,241c72.3,0,111.83-59.9,111.83-111.84,0-1.71,0-3.4-.1-5.09C235.62,118.54,244.84,113.37,250.11,105.48Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_facebook" href="https://www.facebook.com/ncbi.nlm" aria-label="Facebook"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<title>Facebook</title>
<path class="cls-11" d="M210.5,115.12H171.74V97.82c0-8.14,5.39-10,9.19-10h27.14V52l-39.32-.12c-35.66,0-42.42,26.68-42.42,43.77v19.48H99.09v36.32h27.24v109h45.41v-109h35Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_linkedin" href="https://www.linkedin.com/company/ncbinlm" aria-label="LinkedIn"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<title>LinkedIn</title>
<path class="cls-11" d="M101.64,243.37H57.79v-114h43.85Zm-22-131.54h-.26c-13.25,0-21.82-10.36-21.82-21.76,0-11.65,8.84-21.15,22.33-21.15S101.7,78.72,102,90.38C102,101.77,93.4,111.83,79.63,111.83Zm100.93,52.61A17.54,17.54,0,0,0,163,182v61.39H119.18s.51-105.23,0-114H163v13a54.33,54.33,0,0,1,34.54-12.66c26,0,44.39,18.8,44.39,55.29v58.35H198.1V182A17.54,17.54,0,0,0,180.56,164.44Z">
</path>
</svg></a>
<a class="footer-icon" id="footer_github" href="https://github.com/ncbi" aria-label="GitHub"><svg data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 300 300">
<defs>
<style>
.cls-11,
.cls-12 {
fill: #737373;
}
.cls-11 {
fill-rule: evenodd;
}
</style>
</defs>
<title>GitHub</title>
<path class="cls-11" d="M151.36,47.28a105.76,105.76,0,0,0-33.43,206.1c5.28,1,7.22-2.3,7.22-5.09,0-2.52-.09-10.85-.14-19.69-29.42,6.4-35.63-12.48-35.63-12.48-4.81-12.22-11.74-15.47-11.74-15.47-9.59-6.56.73-6.43.73-6.43,10.61.75,16.21,10.9,16.21,10.9,9.43,16.17,24.73,11.49,30.77,8.79,1-6.83,3.69-11.5,6.71-14.14C108.57,197.1,83.88,188,83.88,147.51a40.92,40.92,0,0,1,10.9-28.39c-1.1-2.66-4.72-13.42,1-28,0,0,8.88-2.84,29.09,10.84a100.26,100.26,0,0,1,53,0C198,88.3,206.9,91.14,206.9,91.14c5.76,14.56,2.14,25.32,1,28a40.87,40.87,0,0,1,10.89,28.39c0,40.62-24.74,49.56-48.29,52.18,3.79,3.28,7.17,9.71,7.17,19.58,0,14.15-.12,25.54-.12,29,0,2.82,1.9,6.11,7.26,5.07A105.76,105.76,0,0,0,151.36,47.28Z">
</path>
<path class="cls-12" d="M85.66,199.12c-.23.52-1.06.68-1.81.32s-1.2-1.06-.95-1.59,1.06-.69,1.82-.33,1.21,1.07.94,1.6Zm-1.3-1">
</path>
<path class="cls-12" d="M90,203.89c-.51.47-1.49.25-2.16-.49a1.61,1.61,0,0,1-.31-2.19c.52-.47,1.47-.25,2.17.49s.82,1.72.3,2.19Zm-1-1.08">
</path>
<path class="cls-12" d="M94.12,210c-.65.46-1.71,0-2.37-.91s-.64-2.07,0-2.52,1.7,0,2.36.89.65,2.08,0,2.54Zm0,0"></path>
<path class="cls-12" d="M99.83,215.87c-.58.64-1.82.47-2.72-.41s-1.18-2.06-.6-2.7,1.83-.46,2.74.41,1.2,2.07.58,2.7Zm0,0">
</path>
<path class="cls-12" d="M107.71,219.29c-.26.82-1.45,1.2-2.64.85s-2-1.34-1.74-2.17,1.44-1.23,2.65-.85,2,1.32,1.73,2.17Zm0,0">
</path>
<path class="cls-12" d="M116.36,219.92c0,.87-1,1.59-2.24,1.61s-2.29-.68-2.3-1.54,1-1.59,2.26-1.61,2.28.67,2.28,1.54Zm0,0">
</path>
<path class="cls-12" d="M124.42,218.55c.15.85-.73,1.72-2,1.95s-2.37-.3-2.52-1.14.73-1.75,2-2,2.37.29,2.53,1.16Zm0,0"></path>
</svg></a>
<a class="footer-icon" id="footer_blog" href="https://ncbiinsights.ncbi.nlm.nih.gov/" aria-label="Blog">
<svg id="Layer_1" data-name="Layer 1" xmlns="http://www.w3.org/2000/svg" viewBox="0 0 40 40"><defs><style>.cls-1{fill:#737373;}</style></defs><path class="cls-1" d="M14,30a4,4,0,1,1-4-4,4,4,0,0,1,4,4Zm11,3A19,19,0,0,0,7.05,15a1,1,0,0,0-1,1v3a1,1,0,0,0,.93,1A14,14,0,0,1,20,33.07,1,1,0,0,0,21,34h3a1,1,0,0,0,1-1Zm9,0A28,28,0,0,0,7,6,1,1,0,0,0,6,7v3a1,1,0,0,0,1,1A23,23,0,0,1,29,33a1,1,0,0,0,1,1h3A1,1,0,0,0,34,33Z"></path></svg>
</a>
</div>
</div>
</section>
<section class="container-fluid bg-primary">
<div class="container pt-5">
<div class="row mt-3">
<div class="col-lg-3 col-12">
<p><a class="text-white" href="https://www.nlm.nih.gov/socialmedia/index.html">Connect with NLM</a></p>
<ul class="list-inline social_media">
<li class="list-inline-item"><a href="https://twitter.com/NLM_NIH" aria-label="Twitter" target="_blank" rel="noopener noreferrer"><svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<style type="text/css">
.st20 {
fill: #FFFFFF;
}
.st30 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
</style>
<title>SM-Twitter</title>
<g>
<g>
<g>
<path class="st20" d="M192.9,88.1c-5,2.2-9.2,2.3-13.6,0.1c5.7-3.4,6-5.8,8.1-12.3c-5.4,3.2-11.4,5.5-17.6,6.7
c-10.5-11.2-28.1-11.7-39.2-1.2c-7.2,6.8-10.2,16.9-8,26.5c-22.3-1.1-43.1-11.7-57.2-29C58,91.6,61.8,107.9,74,116
c-4.4-0.1-8.7-1.3-12.6-3.4c0,0.1,0,0.2,0,0.4c0,13.2,9.3,24.6,22.3,27.2c-4.1,1.1-8.4,1.3-12.5,0.5c3.6,11.3,14,19,25.9,19.3
c-11.6,9.1-26.4,13.2-41.1,11.5c12.7,8.1,27.4,12.5,42.5,12.5c51,0,78.9-42.2,78.9-78.9c0-1.2,0-2.4-0.1-3.6
C182.7,97.4,189.2,93.7,192.9,88.1z"></path>
</g>
</g>
<circle class="st30" cx="124.4" cy="128.8" r="108.2"></circle>
</g>
</svg></a></li>
<li class="list-inline-item"><a href="https://www.facebook.com/nationallibraryofmedicine" aria-label="Facebook" rel="noopener noreferrer" target="_blank">
<svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<style type="text/css">
.st10 {
fill: #FFFFFF;
}
.st110 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
</style>
<title>SM-Facebook</title>
<g>
<g>
<path class="st10" d="M159,99.1h-24V88.4c0-5,3.3-6.2,5.7-6.2h16.8V60l-24.4-0.1c-22.1,0-26.2,16.5-26.2,27.1v12.1H90v22.5h16.9
v67.5H135v-67.5h21.7L159,99.1z"></path>
</g>
</g>
<circle class="st110" cx="123.6" cy="123.2" r="108.2"></circle>
</svg>
</a></li>
<li class="list-inline-item"><a href="https://www.youtube.com/user/NLMNIH" aria-label="Youtube" target="_blank" rel="noopener noreferrer"><svg version="1.1" xmlns="http://www.w3.org/2000/svg" xmlns:xlink="http://www.w3.org/1999/xlink" x="0px" y="0px" viewBox="0 0 249 249" style="enable-background:new 0 0 249 249;" xml:space="preserve">
<title>SM-Youtube</title>
<style type="text/css">
.st4 {
fill: none;
stroke: #FFFFFF;
stroke-width: 8;
stroke-miterlimit: 10;
}
.st5 {
fill: #FFFFFF;
}
</style>
<circle class="st4" cx="124.2" cy="123.4" r="108.2"></circle>
<g transform="translate(0,-952.36218)">
<path class="st5" d="M88.4,1037.4c-10.4,0-18.7,8.3-18.7,18.7v40.1c0,10.4,8.3,18.7,18.7,18.7h72.1c10.4,0,18.7-8.3,18.7-18.7
v-40.1c0-10.4-8.3-18.7-18.7-18.7H88.4z M115.2,1058.8l29.4,17.4l-29.4,17.4V1058.8z"></path>
</g>
</svg></a></li>
</ul>
</div>
<div class="col-lg-3 col-12">
<p class="address_footer text-white">National Library of Medicine<br>
<a href="https://www.google.com/maps/place/8600+Rockville+Pike,+Bethesda,+MD+20894/@38.9959508,-77.101021,17z/data=!3m1!4b1!4m5!3m4!1s0x89b7c95e25765ddb:0x19156f88b27635b8!8m2!3d38.9959508!4d-77.0988323" class="text-white" target="_blank" rel="noopener noreferrer">8600 Rockville Pike<br>
Bethesda, MD 20894</a></p>
</div>
<div class="col-lg-3 col-12 centered-lg">
<p><a href="https://www.nlm.nih.gov/web_policies.html" class="text-white">Web Policies</a><br>
<a href="https://www.nih.gov/institutes-nih/nih-office-director/office-communications-public-liaison/freedom-information-act-office" class="text-white">FOIA</a><br>
<a href="https://www.hhs.gov/vulnerability-disclosure-policy/index.html" class="text-white" id="vdp">HHS Vulnerability Disclosure</a></p>
</div>
<div class="col-lg-3 col-12 centered-lg">
<p><a class="supportLink text-white" href="https://support.nlm.nih.gov/">Help</a><br>
<a href="https://www.nlm.nih.gov/accessibility.html" class="text-white">Accessibility</a><br>
<a href="https://www.nlm.nih.gov/careers/careers.html" class="text-white">Careers</a></p>
</div>
</div>
<div class="row">
<div class="col-lg-12 centered-lg">
<nav class="bottom-links">
<ul class="mt-3">
<li>
<a class="text-white" href="//www.nlm.nih.gov/">NLM</a>
</li>
<li>
<a class="text-white" href="https://www.nih.gov/">NIH</a>
</li>
<li>
<a class="text-white" href="https://www.hhs.gov/">HHS</a>
</li>
<li>
<a class="text-white" href="https://www.usa.gov/">USA.gov</a>
</li>
</ul>
</nav>
</div>
</div>
</div>
</section>
</footer>
<script type="text/javascript" src="js/nwds.js"></script>
<script type="text/javascript" src="js/ncbipopup.js"></script>
<script type="text/javascript" src="js/remote_data_provider.js?v=2"></script>
<script type="text/javascript" src="js/headerNew.js?v=2"></script>
<script src="js/uswds.min.js"></script>
<script type="text/javascript" src="/portal/portal3rc.fcgi/rlib/js/InstrumentOmnitureBaseJS/InstrumentNCBIBaseJS/InstrumentPageStarterJS.js"></script>
<!--
<script type="text/javascript" src="://www.ncbi.nlm.nih.gov/portal/portal3rc.fcgi/supportedbrowsers/js/nonportalbc_min.js"></script>
<script type="text/javascript">$("#browsers_ajax").browserCheck();</script>
-->
</div><!--/#wrap-->
</body>
</html>
Exercise: BLAST IT!¶
Use the BLAST URLAPI to find structures with similar sequence to a user-supplied FASTA protein sequence. Your script will take the name of a FASTA file as its only argument. The contents of this file should be provided as the QUERY parameter in a BLAST URL PUT request querying the pdb database (DATABASE=pdb) using blastp (PROGRAM=blastp). You should extract the RID from the response using a regular expression.
Using the RID, you then submit a GET request. Your GET request may either return the desired data, or it may return a status HTML page (even if you request XML) if the request hasn't finished. You should look for the presence of the string Status=WAITING in the response. If this string is present, you should repeat your GET request every 5 seconds (time.sleep(5)) until you get a response without it. It is typical for it to take 30 seconds.
Print out the final XML response.
Example fasta file: http://mscbio2025.net/files/brca.fasta
