%%html
<script src="https://bits.csb.pitt.edu/preamble.js"></script>
Dictionary¶
A dictionary is an unordered collection of key,value pairs where the keys are mapped to values.
A dictionary object is indexed by the key to get the value.
d = dict()
d['key'] = 'value'
d[0] = ['x','y','z']
print(d)
{'key': 'value', 0: ['x', 'y', 'z']}
Initializing dicts¶
empty = dict()
alsoempty = {}
Specify key:value pairs within curly braces.
example = {'a': 1, 'b':2}
example
{'a': 1, 'b': 2}
Add new values by indexing with new/existing key
example['a'] = 0
example['z'] = 26
example
{'a': 0, 'b': 2, 'z': 26}
Accessing values¶
example['c'] #keys must exist
--------------------------------------------------------------------------- KeyError Traceback (most recent call last) Cell In[6], line 1 ----> 1 example['c'] #keys must exist KeyError: 'c'
Use in to test for membership
'c' in example
False
if 'c' not in example:
example['c'] = 0
example
{'a': 0, 'b': 2, 'z': 26, 'c': 0}
Methods¶
example.keys()
dict_keys(['a', 'b', 'z', 'c'])
example.values()
dict_values([0, 2, 26, 0])
example.items()
dict_items([('a', 0), ('b', 2), ('z', 26), ('c', 0)])
def count(vals):
cnts = {}
for x in vals:
cnts[x] += 1
return cnts
d = count(['a','a','b','a','c','b'])
%%html
<div id="dictdefault" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#dictdefault';
jQuery(divid).asker({
id: divid,
question: "What is the value of <tt>d['a']</tt>?",
answers: ["0","1","2","3","6",'Error'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
The Fix¶
def count(vals):
cnts = {}
for x in vals:
if x not in cnts:
cnts[x] = 0
cnts[x] += 1
return cnts
d = count(['a','a','b','a','c','b'])
d['a']
3
If this seems annoying, checkout collections.defaultdict
Sets¶
A set is an unordered collection with no duplicate elements. Basic uses include membership testing and eliminating duplicate entries. Set objects also support mathematical operations like union, intersection, difference, and symmetric difference.
Can initialize with a list
stuff = set(['a','b','a','d','x','a','e'])
stuff
{'a', 'b', 'd', 'e', 'x'}
Sets are not indexed - use add to insert new elements.
stuff.add('y')
set operations¶
Efficient membership testing
'y' in stuff
True
stuff2 = set(['a','b','c'])
print('and',stuff & stuff2) #intersection
and {'a', 'b'}
print('or', stuff | stuff2)
or {'e', 'x', 'd', 'c', 'b', 'a', 'y'}
print('diff', stuff - stuff2)
diff {'e', 'd', 'x', 'y'}
s = set([1,2,2,3,3,3,4,4,4,4])
%%html
<div id="dictset" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#dictset';
jQuery(divid).asker({
id: divid,
question: "What is <tt>len(s)</tt>?",
answers: ["0","1","3","4","9","10",'Error'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
a = set([1,2,2,3])
b = set([2,3,3,4])
c = a & b
%%html
<div id="setinter" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#setinter';
jQuery(divid).asker({
id: divid,
question: "What is <tt>c</tt>?",
answers: ["{1,2,3,4}","{2,2,3,3}","{2,3}","{1,4}",'Error'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Tuples¶
A tuple is an immutable list.
t = tuple([1,2,3])
t
(1, 2, 3)
Tuples are initialized the same way as lists, just with parentheses
t = ('x',0,3.0)
l = ['x',0,3.0]
t,l
(('x', 0, 3.0), ['x', 0, 3.0])
'%s %d' % ('hello',3) # second operand of string % operator is tuple
'hello 3'
t = ('x',0,3.0)
t[2] += 1
%%html
<div id="dictimm" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#dictimm';
jQuery(divid).asker({
id: divid,
question: "What is the value of <tt>t[2]</tt>?",
answers: ["0","1","3.0","4.0",'Error'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Keys¶
The keys of a dictionary or set should be immutable. Examples of immutable types are numbers, strings, tuples (that contain only immutable objects) and frozensets.
example[(1,2)] = 'a'
example[[1,2]] = 'a'
--------------------------------------------------------------------------- TypeError Traceback (most recent call last) Cell In[31], line 1 ----> 1 example[[1,2]] = 'a' TypeError: unhashable type: 'list'
Dictionaries and sets efficiently store data based on the properties of the key - if the properties of the key can change, then the data structure is broken and the data is not where it should be.
Efficiency¶
Imagine we have two lists, l1 and l2, and we want do something with the items that are in common in both lists. Here we'll just count them.
def listcnt(l1, l2):
cnt = 0
for x in l1:
if x in l2:
cnt += 1
return cnt
def setcnt(l1, l2):
cnt = 0
s1 = set(l1)
s2 = set(l2)
for x in s1:
if x in s2:
cnt += 1
return cnt
These two functions generate the same answer if the lists have all distinct elements.
%%html
<div id="dictspeed" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#dictspeed';
jQuery(divid).asker({
id: divid,
question: "Which function is faster?",
answers: ['listcnt, by a lot','listcnt, by a little','about the same','setcnt, by a little','setcnt, by a lot'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
import time
l1 = list(range(40000))
l2 = list(range(1000,10000))
t0 = time.time()
listcnt(l1,l2)
t1 = time.time()
setcnt(l1,l2)
t2 = time.time()
print("listcnt time: ",t1-t0,'\nsetcnt time:',t2-t1)
listcnt time: 2.0174601078033447 setcnt time: 0.0019466876983642578
Can you think of another way?
t0 = time.time()
len(set(l1) & set(l2))
t3 = time.time()
print("set intersection time:",t3-t0)
set intersection time: 0.006762027740478516
Do not do membership testing on lists¶
- Lists are ordered by the programmer, which means Python must examine every element to determine (non)membership.
- Dictionaries and sets are unordered so Python can store the contents in a way that makes membership testing efficient (using hashing)
Hashing¶
The keys of sets and dictionaries must be hashable types. Technically, this means they define methods __eq__ (or __cmp__) and __hash__.
A hash function (__hash__) takes an arbitrary object and produces a number. Objects that are identical (according to __eq__ or __cmp__) must hash to the same hash value.
What does this get us?¶
Accessing an element of an array of elements in memory (random access...) is as simple as computing a memory address (base of array plus offset). This is great if our keys are a dense array of integers (then we have a lookup table).
Hash functions provide a way to quickly index into an array of data even when are keys are arbitray objects.
Hashing¶
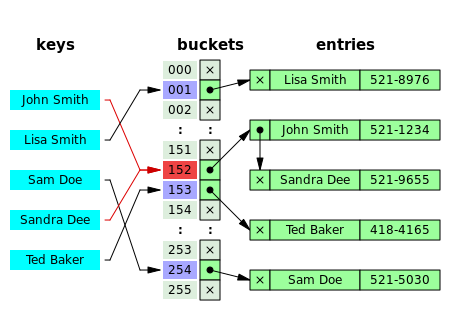
In this figure (from Wikipedia), collisions are resolved through chaining.
With a good hash function and enough buckets, no chain (list) will be more than a few elements long and accessing our data will be constant time (on average).
The position of an element in the table is determined by its hash value and the table size.
position = obj.__hash__() % table_size
Hash Functions¶
A good hash function of an object has the following properties:
- The hash value is fully determined by the object (deterministic).
- The hash function uses all the object's data (if relevant).
- The hash function is uniform (evenly distributes keys across the range)
The range of the hash function is usual the integers ($2^{32}$ or $2^{64}$).
print(hash(3), hash(1435080909832), hash('cat'), hash((3,'cat')))
3 1435080909832 8944782290083313456 2804537357584593689
Implementing a Hash Function¶
You only need to implement a hash function when defining your own object types (which we haven't talked about yet...)
The best (and easiest) approach is to combine the built-in hash functions on your data.
a = 'cat'
b = 'dog'
print(hash(a))
print(hash(b))
print(hash( (a,b) ))
8944782290083313456 -6500199754053014813 -5791914484131828174
Takeaway¶
sets, frozensets, and dicts use hashing to provide extremely efficient membership testing and (for dicts) value lookup.
However, you cannot rely on the order of data in these data structures. In fact, the order of items will change as you add and delete.
s = set([3,999])
s
{3, 999}
s.add(1000)
print(s)
s.update([1001,1002,1003])
{1000, 3, 999}
print(s)
{3, 999, 1000, 1001, 1002, 1003}
polyfit¶
Takes $x$ values, $y$ values, and degree of polynomial. Returns coefficients with least squares fit (highest degree first).
%%html
<div id="polydeg" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#polydeg';
jQuery(divid).asker({
id: divid,
question: "What degree is the polynomial 4x^2 + 5x - 3?",
answers: ['1','2','3','4','5'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
import numpy as np
xvals = np.linspace(-1,2,20)
yvals = xvals**3 +np.random.random(20) #adds random numbers from 0 to 1 to 20 values of xvals
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(xvals,yvals,'o');
polyfit¶
deg1 = np.polyfit(xvals,yvals,1)
deg2 = np.polyfit(xvals,yvals,2)
deg3 = np.polyfit(xvals,yvals,3)
deg1
array([2.2623292 , 0.60192043])
deg2
array([ 1.39717871, 0.86515049, -0.20697251])
%%html
<div id="deg3" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#deg3';
jQuery(divid).asker({
id: divid,
question: "What is the expected value of deg3[-1]?",
answers: ['-1','0','0.5','1'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
%%html
<div id="deg3_2" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="https://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#deg3_2';
jQuery(divid).asker({
id: divid,
question: "What is the expected value of deg3[0]?",
answers: ['-1','0','0.5','1'],
server: "https://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
deg3
array([ 1.01783337, -0.12957134, 0.11488606, 0.42261805])
deg3[-1]
0.4226180479244404
poly1d¶
Construct a polynomial function from coefficients
p1 = np.poly1d(deg1)
p2 = np.poly1d(deg2)
p3 = np.poly1d(deg3)
p1(2),p2(2),p3(2)
(5.126578820886959, 7.112043304060662, 8.2767717367996)
p1(0),p2(0),p3(0)
(0.6019204286737281, -0.20697250891555835, 0.4226180479244404)
plt.plot(xvals,yvals,'o',xvals,p1(xvals),'-',xvals,p2(xvals),'-',xvals,p3(xvals),'-');
scipy.optimize.curve_fit¶
Optimize fit to an arbitrary function. Returns optimal values (for least squares) of parameters along with covariance estimate.
Provide python function that takes x value and any parameters.
from scipy.optimize import curve_fit
def tanh(x,a,b):
return b*np.tanh(a+x)
popt,pconv = curve_fit(tanh, xvals, yvals)
popt
array([0.24845812, 3.44033153])
plt.plot(xvals,yvals,'o',xvals,popt[1]*np.tanh(popt[0]+xvals)); plt.show()
Let's Analyze Data!¶
!wget https://MSCBIO2025.github.io/files/kd
!wget https://MSCBIO2025.github.io/files/aff.min
!wget https://MSCBIO2025.github.io/files/aff.score
!head kd
set1_1 3.34 set1_100 6.3 set1_102 8.62 set1_103 6.44 set1_104 -0.15 set1_105 8.48 set1_106 7.01 set1_107 4.61 set1_108 7.55 set1_109 7.96
kd contains experimental values while aff.min and aff.score contain computational predictions of the experimental values. Each line has a name and value.
How good are the predictions?
How do we want to load and store the data?¶
The rows of the provided files are not in the same order.
We only care about points that are in all three files.
Hint: use dictionary to associate names with values.
import numpy as np
def makedict(fname):
f = open(fname)
retdict = {}
for line in f:
(name,value) = line.split()
retdict[name] = float(value)
return retdict
kdvalues = makedict('kd')
scorevalues = makedict('aff.score')
minvalues = makedict('aff.min')
names = []
kdlist = []
scorelist = []
minlist = []
for name in sorted(kdvalues.keys()):
if name in scorevalues and name in minvalues:
names.append(name)
kdlist.append(kdvalues[name])
scorelist.append(scorevalues[name])
minlist.append(minvalues[name])
kds = np.array(kdlist)
scores = np.array(scorelist)
mins = np.array(minlist)
How do we want to visualize the data?¶
Plot experiment value vs. predicted values (two series, scatterplot).
%matplotlib inline
import matplotlib.pylab as plt
plt.plot(kds,scores,'o',alpha=0.5,label='score')
plt.plot(kds,mins,'o',alpha=0.5,label='min')
plt.legend(numpoints=1)
plt.xlim(0,18)
plt.ylim(0,18)
plt.xlabel('Experiment')
plt.ylabel('Prediction')
plt.gca().set_aspect('equal')
plt.show()
Aside: Visualizing dense 2D distributions¶
seaborn - extends matplotlib to make some hard things easy
import seaborn as sns # package that sits on top of matplotlib
sns.jointplot(x=kds, y=scores);
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
sns.jointplot(x=kds, y=scores, kind='hex');
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
sns.jointplot(x=kds, y=scores, kind='kde');
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1498: FutureWarning: is_categorical_dtype is deprecated and will be removed in a future version. Use isinstance(dtype, CategoricalDtype) instead
if pd.api.types.is_categorical_dtype(vector):
/opt/homebrew/Caskroom/miniforge/base/lib/python3.10/site-packages/seaborn/_oldcore.py:1119: FutureWarning: use_inf_as_na option is deprecated and will be removed in a future version. Convert inf values to NaN before operating instead.
with pd.option_context('mode.use_inf_as_na', True):
What is the error?¶
Average absolute? Mean squared?
print("Scores absolute average error:",np.mean(np.abs(scores-kds)))
print("Mins absolute average error:",np.mean(np.abs(mins-kds)))
Scores absolute average error: 2.3590879883381928 Mins absolute average error: 2.682967900874636
print("Scores Mean squared error:",np.mean(np.square(scores-kds)))
print("Mins Mean squared error:",np.mean(np.square(mins-kds)))
Scores Mean squared error: 8.165401751357434 Mins Mean squared error: 10.058501106196793
plt.hist(np.square(kds-mins),25,(0,25),density=True)
plt.show()
ave = np.mean(kds)
print("Average experimental value",ave)
print("Error of predicting the average",np.mean(np.square(kds-ave)))
Average experimental value 6.154081632653062 Error of predicting the average 4.956176343190337
Do the predictions correlate with the observed values?¶
Compute correlations: np.corrcoef, scipy.stats.pearsonr, scipy.stats.spearmanr, scipy.stats.kendalltau
np.corrcoef(kds,scores)
array([[1. , 0.58006564],
[0.58006564, 1. ]])
np.corrcoef(kds,mins)
array([[1. , 0.59026701],
[0.59026701, 1. ]])
import scipy.stats as stats
stats.pearsonr(kds,scores)
PearsonRResult(statistic=0.5800656427326171, pvalue=3.1321668954855117e-32)
stats.spearmanr(kds,scores)
SignificanceResult(statistic=0.5843897097494348, pvalue=8.484372310278012e-33)
stats.kendalltau(kds,scores)
SignificanceResult(statistic=0.4128988002305019, pvalue=4.1227137153591064e-30)
What is the linear relationship?¶
fit = np.polyfit(kds,scores,1)
fit
array([0.62828046, 4.19091432])
line = np.poly1d(fit) #converts coefficients into function
line(3)
6.075755700166205
xpoints = np.linspace(0,18,100) #make 100 xcoords
plt.plot(kds,scores,'o',alpha=0.5,label='score')
plt.plot(kds,mins,'o',alpha=0.5,label='min')
plt.xlim(0,18)
plt.ylim(0,18)
plt.xlabel('Experiment')
plt.ylabel('Prediction')
plt.gca().set_aspect('equal')
plt.plot(xpoints,xpoints,'k')
plt.plot(xpoints,line(xpoints),label="fit",linewidth=2)
plt.legend(loc='lower right')
plt.show()
What happens if we rescale the predictions?¶
Apply the linear fit to the predicted values.
f2 = np.polyfit(scores,kds,1)
print("Fit:",f2)
fscores = scores*f2[0]+f2[1]
print("Scores Mean squared error:",np.mean(np.square(scores-kds)))
print("Fit Scores Mean squared error:",np.mean(np.square(fscores-kds)))
Fit: [0.53555088 1.83893211] Scores Mean squared error: 8.165401751357434 Fit Scores Mean squared error: 3.2885412091132413
plt.plot(kds,scores,'o',alpha=0.5,label='score')
plt.plot(kds,fscores,'o',alpha=0.5,label='fit')
plt.xlim(0,18)
plt.ylim(0,18)
plt.xlabel('Experiment')
plt.ylabel('Prediction')
plt.gca().set_aspect('equal')
plt.plot(xpoints,xpoints)
plt.legend(loc='lower right')
plt.show()
stats.pearsonr(kds,fscores)
PearsonRResult(statistic=0.5800656427326171, pvalue=3.1321668954855117e-32)