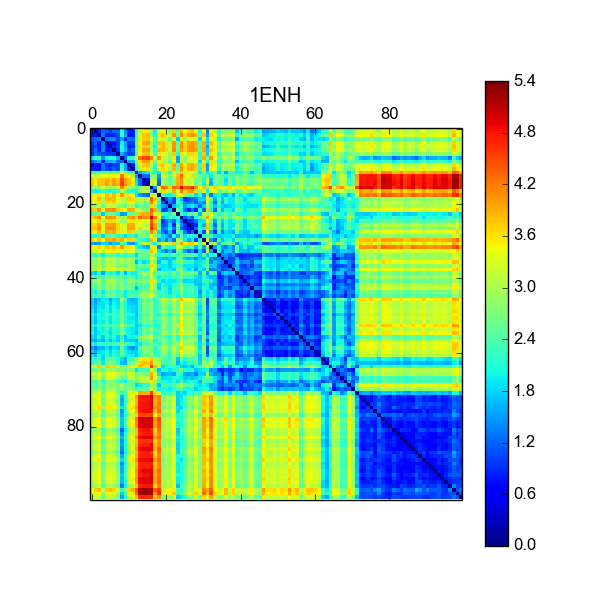
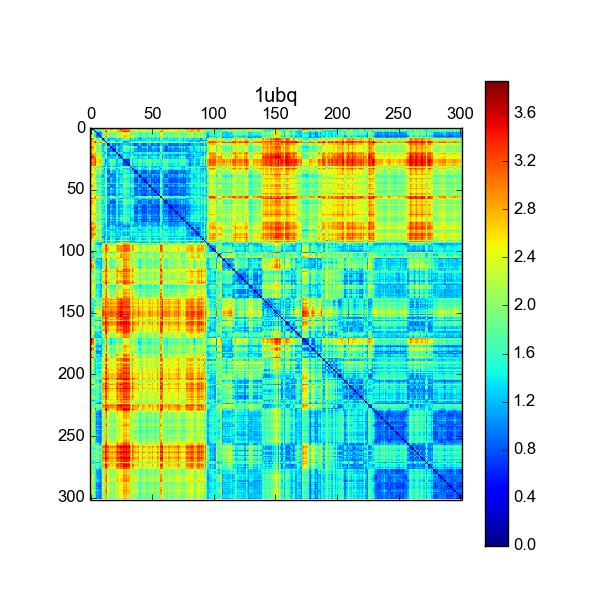
MDAnalysis Review¶
In [2]:
import MDAnalysis
from MDAnalysis.analysis.rms import * #this pulls in an rmsd function
universe = MDAnalysis.Universe('shmt2.prmtop', 'shmt2.dcd')
prot = universe.select_atoms('protein')
startref = prot.positions
universe.trajectory[-1]
endref = prot.positions
startrmsd = []
endrmsd = []
for ts in universe.trajectory:
startrmsd.append(rmsd(startref,prot.positions))
endrmsd.append(rmsd(endref,prot.positions))
In [3]:
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(startrmsd,label='start')
plt.plot(endrmsd,label='end')
plt.legend(loc='lower center')
plt.ylabel('RMSD')
plt.xlabel("Frame");
MDAnalysis.analysis.contacts¶
Analysis of native contacts q over a trajectory.
- a “contact” exists between two atoms i and j if the distance between them is smaller than a given radius
- a “native contact” exists between i and j if a contact exists and if the contact also exists between the equivalent atoms in a reference structure or conformation
What counts as a contact?
- Hard Cut: To count as a contact the atoms i and j have to be at least as close as in the reference structure.
- Soft Cut: The atom pair i and j is assigned based on a soft potential ($\lambda = 1.8$ and $\beta = 5.0$ by default) $$Q(r, r_0) = \frac{1}{1 + e^{\beta (r - \lambda r_0)}}$$
- Radius Cut: To count as a contact the atoms i and j cannot be further apart then some distance radius.
In [4]:
x = np.arange(0.0, 6.0, 0.01)
y = 1.0/(1.0 + np.exp(5.0*(x-1.8*2)))
plt.axvline(2,color='grey',linewidth=1)
plt.plot(x,y);
In [5]:
from MDAnalysis.analysis.contacts import *
help(MDAnalysis.analysis.contacts.Contacts)
Help on class Contacts in module MDAnalysis.analysis.contacts: class Contacts(MDAnalysis.analysis.base.AnalysisBase) | Contacts(u, select, refgroup, method='hard_cut', radius=4.5, pbc=True, kwargs=None, **basekwargs) | | Calculate contacts based observables. | | The standard methods used in this class calculate the fraction of native | contacts *Q* from a trajectory. | | | .. rubric:: Contact API | | By defining your own method it is possible to calculate other observables | that only depend on the distances and a possible reference distance. The | **Contact API** prescribes that this method must be a function with call | signature ``func(r, r0, **kwargs)`` and must be provided in the keyword | argument `method`. | | Attributes | ---------- | results.timeseries : numpy.ndarray | 2D array containing *Q* for all refgroup pairs and analyzed frames | | timeseries : numpy.ndarray | Alias to the :attr:`results.timeseries` attribute. | | .. deprecated:: 2.0.0 | Will be removed in MDAnalysis 3.0.0. Please use | :attr:`results.timeseries` instead. | | | .. versionchanged:: 1.0.0 | ``save()`` method has been removed. Use ``np.savetxt()`` on | :attr:`Contacts.results.timeseries` instead. | .. versionchanged:: 1.0.0 | added ``pbc`` attribute to calculate distances using PBC. | .. versionchanged:: 2.0.0 | :attr:`timeseries` results are now stored in a | :class:`MDAnalysis.analysis.base.Results` instance. | .. versionchanged:: 2.2.0 | :class:`Contacts` accepts both AtomGroup and string for `select` | | Method resolution order: | Contacts | MDAnalysis.analysis.base.AnalysisBase | builtins.object | | Methods defined here: | | __init__(self, u, select, refgroup, method='hard_cut', radius=4.5, pbc=True, kwargs=None, **basekwargs) | Parameters | ---------- | u : Universe | trajectory | select : tuple(AtomGroup, AtomGroup) | tuple(string, string) | two contacting groups that change over time | refgroup : tuple(AtomGroup, AtomGroup) | two contacting atomgroups in their reference conformation. This | can also be a list of tuples containing different atom groups | radius : float, optional (4.5 Angstroms) | radius within which contacts exist in refgroup | method : string | callable (optional) | Can either be one of ``['hard_cut' , 'soft_cut', 'radius_cut']`` or a callable | with call signature ``func(r, r0, **kwargs)`` (the "Contacts API"). | pbc : bool (optional) | Uses periodic boundary conditions to calculate distances if set to ``True``; the | default is ``True``. | kwargs : dict, optional | dictionary of additional kwargs passed to `method`. Check | respective functions for reasonable values. | verbose : bool (optional) | Show detailed progress of the calculation if set to ``True``; the | default is ``False``. | | Notes | ----- | | .. versionchanged:: 1.0.0 | Changed `selection` keyword to `select` | | ---------------------------------------------------------------------- | Readonly properties defined here: | | timeseries | | ---------------------------------------------------------------------- | Methods inherited from MDAnalysis.analysis.base.AnalysisBase: | | run(self, start=None, stop=None, step=None, frames=None, verbose=None) | Perform the calculation | | Parameters | ---------- | start : int, optional | start frame of analysis | stop : int, optional | stop frame of analysis | step : int, optional | number of frames to skip between each analysed frame | frames : array_like, optional | array of integers or booleans to slice trajectory; `frames` can | only be used *instead* of `start`, `stop`, and `step`. Setting | *both* `frames` and at least one of `start`, `stop`, `step` to a | non-default value will raise a :exc:`ValueError`. | | .. versionadded:: 2.2.0 | | verbose : bool, optional | Turn on verbosity | | | .. versionchanged:: 2.2.0 | Added ability to analyze arbitrary frames by passing a list of | frame indices in the `frames` keyword argument. | | ---------------------------------------------------------------------- | Data descriptors inherited from MDAnalysis.analysis.base.AnalysisBase: | | __dict__ | dictionary for instance variables (if defined) | | __weakref__ | list of weak references to the object (if defined)
In [6]:
import MDAnalysis
from MDAnalysis.analysis.contacts import *
u = MDAnalysis.Universe('shmt2.prmtop','shmt2.dcd')
sel_basic = "(resname ARG or resname LYS) and (name NH* or name NZ)"
sel_acidic = "(resname ASP or resname GLU) and (name OE* or name OD*)"
acidic = u.select_atoms(sel_acidic)
basic = u.select_atoms(sel_basic)
CA = Contacts(u, select=(sel_acidic, sel_basic), refgroup=(acidic, basic))
CA.run()
plt.plot(CA.results.timeseries[:, 0], CA.results.timeseries[:, 1])
plt.ylim(0,1)
plt.xlabel("Frame"); plt.ylabel("Fraction native contacts");
In [7]:
CAsoft = Contacts(u, select=(sel_acidic, sel_basic), refgroup=(acidic, basic),method='soft_cut')
CAsoft.run()
plt.plot(CAsoft.results.timeseries[:, 0], CAsoft.results.timeseries[:, 1],label="soft")
plt.plot(CA.results.timeseries[:, 0], CA.results.timeseries[:, 1],label="hard")
plt.legend(loc='best')
plt.ylim(0,1)
plt.xlabel("Frame"); plt.ylabel("Fraction native contacts");
MDAnalysis.analysis.distances¶
Analysis of distances between groups of atoms. Precursor to contact analysis.
In [8]:
from MDAnalysis.analysis.distances import *
distarr = distance_array(acidic.positions,basic.positions)
In [9]:
import MDAnalysis.analysis.distances
help(MDAnalysis.analysis.distances.distance_array)
Help on function distance_array in module MDAnalysis.lib.distances:
distance_array(reference: Union[numpy.ndarray, ForwardRef('AtomGroup')], configuration: Union[numpy.ndarray, ForwardRef('AtomGroup')], box: Optional[numpy.ndarray] = None, result: Optional[numpy.ndarray] = None, backend: str = 'serial') -> numpy.ndarray
Calculate all possible distances between a reference set and another
configuration.
If there are ``n`` positions in `reference` and ``m`` positions in
`configuration`, a distance array of shape ``(n, m)`` will be computed.
If the optional argument `box` is supplied, the minimum image convention is
applied when calculating distances. Either orthogonal or triclinic boxes are
supported.
If a 2D numpy array of dtype ``numpy.float64`` with the shape ``(n, m)``
is provided in `result`, then this preallocated array is filled. This can
speed up calculations.
Parameters
----------
reference :numpy.ndarray or :class:`~MDAnalysis.core.groups.AtomGroup`
Reference coordinate array of shape ``(3,)`` or ``(n, 3)`` (dtype is
arbitrary, will be converted to ``numpy.float32`` internally). Also
accepts an :class:`~MDAnalysis.core.groups.AtomGroup`.
configuration : numpy.ndarray or :class:`~MDAnalysis.core.groups.AtomGroup`
Configuration coordinate array of shape ``(3,)`` or ``(m, 3)`` (dtype is
arbitrary, will be converted to ``numpy.float32`` internally). Also
accepts an :class:`~MDAnalysis.core.groups.AtomGroup`.
box : array_like, optional
The unitcell dimensions of the system, which can be orthogonal or
triclinic and must be provided in the same format as returned by
:attr:`MDAnalysis.coordinates.timestep.Timestep.dimensions`:
``[lx, ly, lz, alpha, beta, gamma]``.
result : numpy.ndarray, optional
Preallocated result array which must have the shape ``(n, m)`` and dtype
``numpy.float64``.
Avoids creating the array which saves time when the function
is called repeatedly.
backend : {'serial', 'OpenMP'}, optional
Keyword selecting the type of acceleration.
Returns
-------
d : numpy.ndarray (``dtype=numpy.float64``, ``shape=(n, m)``)
Array containing the distances ``d[i,j]`` between reference coordinates
``i`` and configuration coordinates ``j``.
.. versionchanged:: 0.13.0
Added *backend* keyword.
.. versionchanged:: 0.19.0
Internal dtype conversion of input coordinates to ``numpy.float32``.
Now also accepts single coordinates as input.
.. versionchanged:: 2.3.0
Can now accept an :class:`~MDAnalysis.core.groups.AtomGroup` as an
argument in any position and checks inputs using type hinting.
In [10]:
plt.matshow(distarr)
plt.colorbar()
plt.xlabel('Basic'); plt.ylabel('Acidic');
In [11]:
%%html
<div id="mdmatrix" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="http://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#mdmatrix';
jQuery(divid).asker({
id: divid,
question: "The previous matrix is?",
answers: ["Gaussian",'Symmetric','Diagonal',
'All of the above','None of the above'],
server: "http://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Distance Analysis¶
Really only interested in short distances
In [12]:
origdistarr = distance_array(acidic.positions,basic.positions)
origdistarr[origdistarr > 6] = 0
plt.matshow(origdistarr)
plt.colorbar()
plt.xlabel('Basic'); plt.ylabel('Acidic');
Analyzing across the trajectory¶
In [13]:
distarr = np.zeros((acidic.n_atoms, basic.n_atoms))
for ts in u.trajectory:
distarr += distance_array(acidic.positions,basic.positions)
distarr /= universe.trajectory.n_frames
In [14]:
distarr[distarr > 6] = 0
plt.matshow(distarr)
plt.colorbar()
plt.xlabel('Basic'); plt.ylabel('Acidic');
In [15]:
%%html
<div id="dmat" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="http://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#dmat';
jQuery(divid).asker({
id: divid,
question: "What are the elements of this matrix?",
answers: ["Max distance",'Final Distance','Average Distance',
'Minimum distance'],
server: "http://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
In [16]:
diff = origdistarr - distarr
plt.matshow(diff,cmap='seismic')
clb = plt.colorbar()
clb.set_label('')
plt.xlabel('Basic'); plt.ylabel('Acidic');
I'm most interested in the loop from residue 368 to 383 (resid 368:383).
How is this loop changing over the course of the simulation (RMSD to first)?
What is happening with the number of hydrogen bonds it is making with the protein?
In [17]:
u.trajectory[0]
loop = u.select_atoms('resid 368:383')
ref = loop.positions
rmsds = []
for ts in u.trajectory:
rmsds.append(rmsd(ref,loop.positions))
plt.plot(rmsds);
Hydrogen Bond analysis¶
In [18]:
from MDAnalysis.analysis.hydrogenbonds import HydrogenBondAnalysis
h = HydrogenBondAnalysis(u,'resid 368:383','protein')
h.run();
In [19]:
counts = h.count_by_time()
plt.plot(h.frames,h.count_by_time())
plt.xlabel("Frame #"); plt.ylabel("HBond Count");
In [20]:
%%html
<div id="hbond1" style="width: 500px"></div>
<script>
$('head').append('<link rel="stylesheet" href="http://bits.csb.pitt.edu/asker.js/themes/asker.default.css" />');
var divid = '#hbond1';
jQuery(divid).asker({
id: divid,
question: "Hydrogen bonds are identified based on?",
answers: ["Atom types",'Distances','Angles',
'All of the above'],
server: "http://bits.csb.pitt.edu/asker.js/example/asker.cgi",
charter: chartmaker})
$(".jp-InputArea .o:contains(html)").closest('.jp-InputArea').hide();
</script>
Actually, what I'm really interested in is comparing SHMT2 and SHMT1...
In [21]:
!wget https://asinansaglam.github.io/python_bio_2022/files/shmt1.prmtop
!wget https://asinansaglam.github.io/python_bio_2022/files/shmt1.dcd
--2022-10-11 08:26:38-- https://asinansaglam.github.io/python_bio_2022/files/shmt1.prmtop Resolving asinansaglam.github.io (asinansaglam.github.io)... 2606:50c0:8000::153, 2606:50c0:8001::153, 2606:50c0:8002::153, ... Connecting to asinansaglam.github.io (asinansaglam.github.io)|2606:50c0:8000::153|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 16909535 (16M) [application/octet-stream] Saving to: ‘shmt1.prmtop’ shmt1.prmtop 100%[===================>] 16.13M 32.2MB/s in 0.5s 2022-10-11 08:26:38 (32.2 MB/s) - ‘shmt1.prmtop’ saved [16909535/16909535] --2022-10-11 08:26:39-- https://asinansaglam.github.io/python_bio_2022/files/shmt1.dcd Resolving asinansaglam.github.io (asinansaglam.github.io)... 2606:50c0:8003::153, 2606:50c0:8002::153, 2606:50c0:8001::153, ... Connecting to asinansaglam.github.io (asinansaglam.github.io)|2606:50c0:8003::153|:443... connected. HTTP request sent, awaiting response... 200 OK Length: 97842196 (93M) [application/octet-stream] Saving to: ‘shmt1.dcd’ shmt1.dcd 100%[===================>] 93.31M 29.3MB/s in 3.3s 2022-10-11 08:26:42 (28.4 MB/s) - ‘shmt1.dcd’ saved [97842196/97842196]
Project¶
Compare the loop regions (resid 368:383 in SHMT2 vs resid 377:392 in SHMT1)
- Compare the backbone RMSDs
- Compare the hydrogen bonding patterns
- Compare the native contacts
In [22]:
shmt1 = MDAnalysis.Universe('shmt1.prmtop', 'shmt1.dcd')
prot1 = shmt1.select_atoms('protein')
In [23]:
prot1.select_atoms('name CA and resid 377:392').resnames
Out[23]:
array(['ASN', 'THR', 'CYS', 'PRO', 'GLY', 'ASP', 'ARG', 'SER', 'ALA',
'LEU', 'ARG', 'PRO', 'SER', 'GLY', 'LEU', 'ARG'], dtype=object)
In [24]:
prot.select_atoms('name CA and resid 368:383').resnames
Out[24]:
array(['ASN', 'THR', 'CYS', 'PRO', 'GLY', 'ASP', 'ARG', 'SER', 'ALA',
'ILE', 'THR', 'PRO', 'GLY', 'GLY', 'LEU', 'ARG'], dtype=object)
In [25]:
import MDAnalysis
shmt2 = MDAnalysis.Universe('shmt2.prmtop','shmt2.dcd')
shmt1 = MDAnalysis.Universe('shmt1.prmtop','shmt1.dcd')
shmt2loopbackbone = shmt2.select_atoms('name CA and resid 368:383')
shmt1loopbackbone = shmt1.select_atoms('name CA and resid 377:392')
ref1 = shmt1loopbackbone.positions
ref2 = shmt2loopbackbone.positions
from MDAnalysis.analysis.rms import rmsd
rmsds1 = []
rmsds2 = []
for ts in shmt1.trajectory:
rmsds1.append(rmsd(ref1,shmt1loopbackbone.positions))
for ts in shmt2.trajectory:
rmsds2.append(rmsd(ref2,shmt2loopbackbone.positions))
import matplotlib.pyplot as plt
%matplotlib inline
plt.plot(rmsds1,label='shmt1',linewidth=3)
plt.plot(rmsds2,'--',label='shmt2',linewidth=3)
plt.xlabel('Frame')
plt.ylabel('RMSD)')
plt.legend(loc='best');
In [26]:
h1 = HydrogenBondAnalysis(shmt1,'resid 377:392','protein')
h1.run();
h2 = HydrogenBondAnalysis(shmt2,'resid 368:383','protein')
h2.run();
plt.plot(h1.count_by_time(),'-',label='shmt1',linewidth=2)
plt.plot(h2.count_by_time(),'--',label='shmt2',linewidth=2)
plt.xlabel('Frame'); plt.ylabel('HBonds')
plt.legend();
In [28]:
loop1 = shmt1.select_atoms('resid 377:392')
prot1 = shmt1.select_atoms('protein')
CA1 = Contacts(shmt1, select=('resid 377:392','protein'), refgroup=(loop1, prot1),method='soft_cut')
CA1.run()
loop2 = shmt2.select_atoms('resid 368:383')
prot2 = shmt2.select_atoms('protein')
CA2 = Contacts(shmt2, select=('resid 368:383','protein'), refgroup=(loop2, prot2),method='soft_cut')
CA2.run()
plt.plot(CA1.results.timeseries[:, 0], CA1.results.timeseries[:, 1],label="shmt1")
plt.plot(CA2.results.timeseries[:, 0], CA2.results.timeseries[:, 1],label="shmt2")
plt.legend(loc='best')
plt.xlabel("Frame"); plt.ylabel("Fraction native contacts");